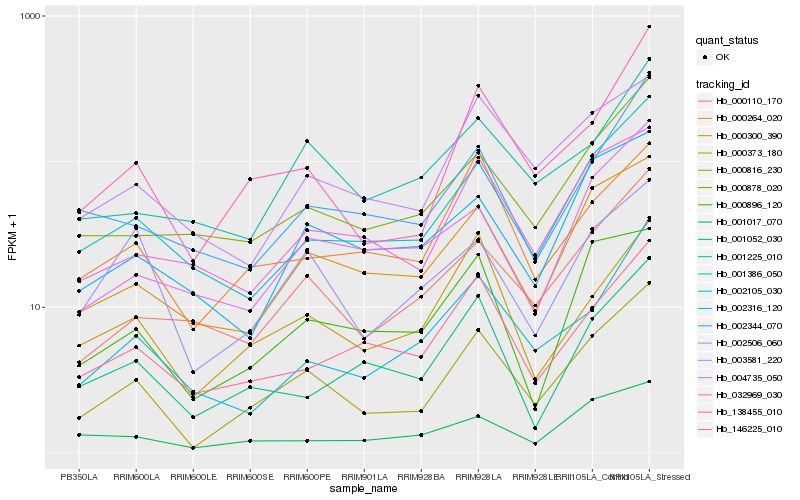
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_230 |
0.0 |
- |
- |
PREDICTED: adenine/guanine permease AZG1 [Jatropha curcas] |
| 2 |
Hb_138455_010 |
0.1180620365 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001386_050 |
0.1361721757 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 4 |
Hb_000300_390 |
0.1384794219 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 5 |
Hb_000110_170 |
0.144644774 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000878_020 |
0.153208138 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 7 |
Hb_146225_010 |
0.1541056391 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 8 |
Hb_003581_220 |
0.1567107336 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000373_180 |
0.157597824 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |
| 10 |
Hb_032969_030 |
0.1577159828 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_002105_030 |
0.1578682041 |
- |
- |
- |
| 12 |
Hb_000264_020 |
0.1621849007 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000896_120 |
0.1629775861 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 14 |
Hb_002344_070 |
0.1637606665 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 15 |
Hb_001017_070 |
0.1643470952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001052_030 |
0.1643606166 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002506_060 |
0.1647533967 |
- |
- |
PREDICTED: probable transporter mch1 [Populus euphratica] |
| 18 |
Hb_001225_010 |
0.1652023118 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_002316_120 |
0.1653115368 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 20 |
Hb_004735_050 |
0.1655656677 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |