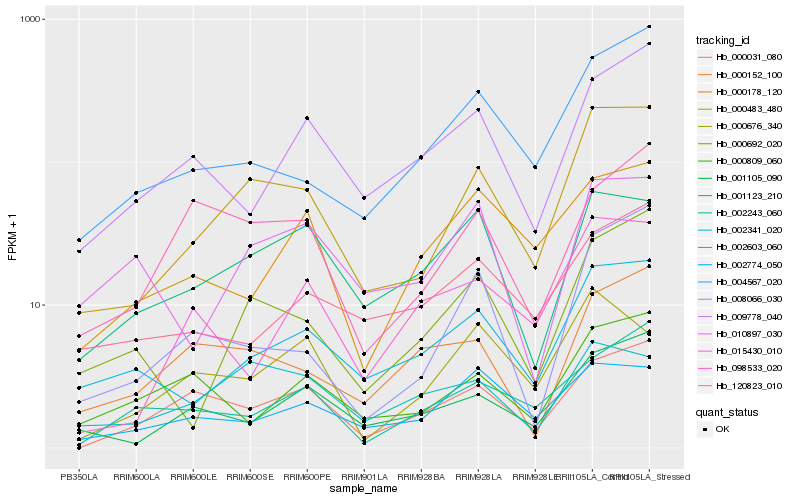
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_480 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002603_060 |
0.158843348 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 3 |
Hb_000692_020 |
0.1646622486 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 4 |
Hb_004567_020 |
0.176380776 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 5 |
Hb_001105_090 |
0.1765709854 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008066_030 |
0.1895057904 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 7 |
Hb_000031_080 |
0.1926749623 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 8 |
Hb_001123_210 |
0.1938444615 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 9 |
Hb_002774_050 |
0.2021875626 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 10 |
Hb_009778_040 |
0.2053714813 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 11 |
Hb_000152_100 |
0.2056728504 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 12 |
Hb_098533_020 |
0.2077899976 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 13 |
Hb_010897_030 |
0.2083474075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002341_020 |
0.2087488069 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 15 |
Hb_120823_010 |
0.2102882457 |
- |
- |
- |
| 16 |
Hb_000809_060 |
0.2137031463 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 17 |
Hb_002243_060 |
0.2156253735 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 18 |
Hb_015430_010 |
0.2161861184 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 19 |
Hb_000676_340 |
0.2185233245 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000178_120 |
0.2192723134 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |