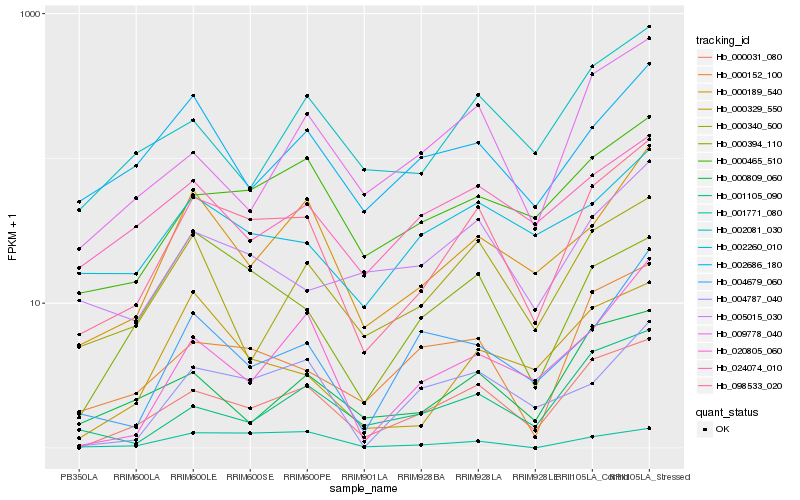
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098533_020 |
0.0 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 2 |
Hb_000031_080 |
0.1286178524 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 3 |
Hb_000189_540 |
0.1551629571 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000465_510 |
0.1659048678 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 5 |
Hb_000152_100 |
0.1664423832 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000340_500 |
0.1802002165 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 7 |
Hb_005015_030 |
0.1827201922 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 8 |
Hb_000394_110 |
0.1830778031 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000809_060 |
0.1863094278 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 10 |
Hb_002081_030 |
0.1866187871 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_002260_010 |
0.1866791319 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 12 |
Hb_004787_040 |
0.1878661788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000329_550 |
0.1887476177 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 14 |
Hb_004679_060 |
0.1896363671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002686_180 |
0.1901916225 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 16 |
Hb_009778_040 |
0.1938788697 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 17 |
Hb_020805_060 |
0.1955731349 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 18 |
Hb_001105_090 |
0.1970959393 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_024074_010 |
0.1984547822 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 20 |
Hb_001771_080 |
0.1995029834 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |