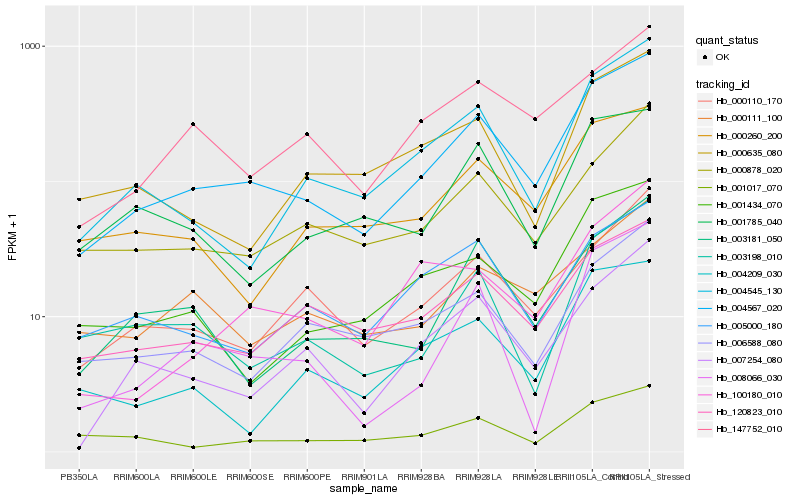
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004567_020 |
0.0 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 2 |
Hb_120823_010 |
0.1285820395 |
- |
- |
- |
| 3 |
Hb_004545_130 |
0.1293865503 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 4 |
Hb_000878_020 |
0.130484877 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 5 |
Hb_000110_170 |
0.1319282595 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_147752_010 |
0.133217316 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 7 |
Hb_007254_080 |
0.1334135472 |
- |
- |
- |
| 8 |
Hb_001434_070 |
0.134476569 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 9 |
Hb_000260_200 |
0.1372407353 |
- |
- |
- |
| 10 |
Hb_003181_050 |
0.1380625041 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 11 |
Hb_006588_080 |
0.1425507436 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 12 |
Hb_003198_010 |
0.1434498634 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |
| 13 |
Hb_005000_180 |
0.1437284434 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 14 |
Hb_000111_100 |
0.1441630356 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 15 |
Hb_004209_030 |
0.1471027739 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001017_070 |
0.1496474409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_100180_010 |
0.1506022605 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 18 |
Hb_008066_030 |
0.1512048252 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 19 |
Hb_000635_080 |
0.1527172111 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 20 |
Hb_001785_040 |
0.1529175167 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |