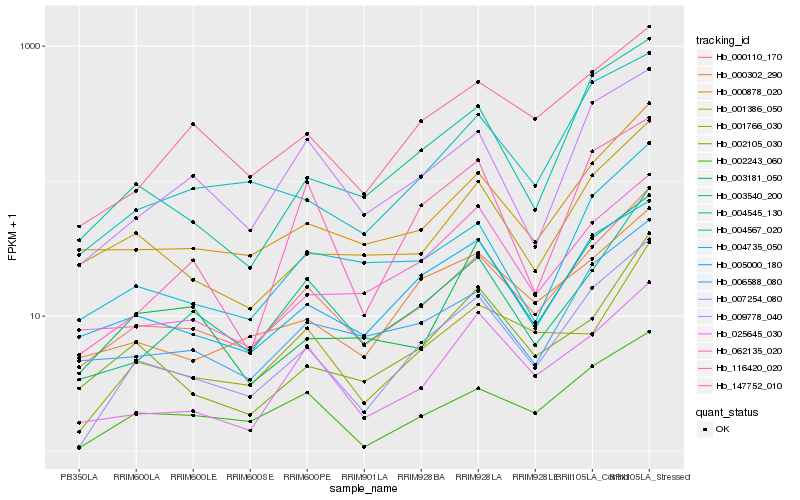
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007254_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_000110_170 |
0.1041879299 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_004545_130 |
0.12245108 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 4 |
Hb_002243_060 |
0.1240551685 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 5 |
Hb_004567_020 |
0.1334135472 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 6 |
Hb_005000_180 |
0.1439038594 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 7 |
Hb_147752_010 |
0.1451252384 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 8 |
Hb_003181_050 |
0.1510664512 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 9 |
Hb_000302_290 |
0.1526502157 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 10 |
Hb_000878_020 |
0.1540173883 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 11 |
Hb_003540_200 |
0.1541275461 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 12 |
Hb_009778_040 |
0.155229964 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 13 |
Hb_004735_050 |
0.1556974709 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006588_080 |
0.1576531103 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 15 |
Hb_001386_050 |
0.1580377163 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 16 |
Hb_116420_020 |
0.1581127452 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 17 |
Hb_001766_030 |
0.1582237074 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 18 |
Hb_002105_030 |
0.1586063006 |
- |
- |
- |
| 19 |
Hb_062135_020 |
0.1591098114 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 20 |
Hb_025645_030 |
0.1599964169 |
- |
- |
- |