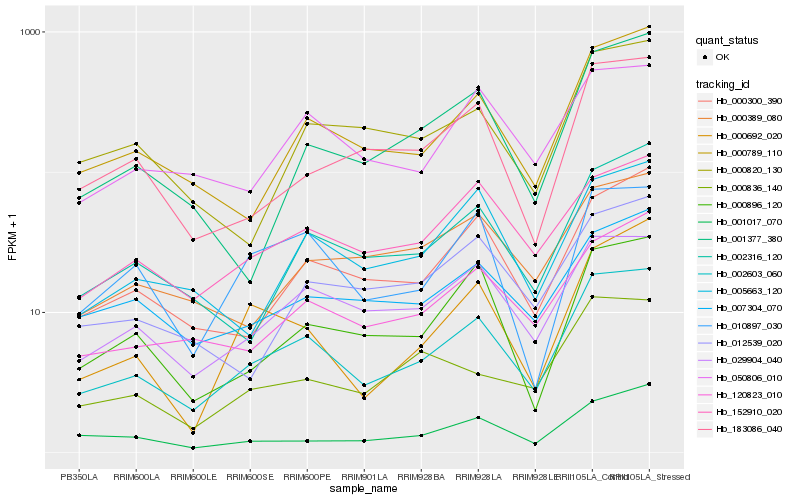
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002603_060 |
0.0 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 2 |
Hb_029904_040 |
0.1027359261 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 3 |
Hb_000789_110 |
0.11312997 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 4 |
Hb_120823_010 |
0.1147302369 |
- |
- |
- |
| 5 |
Hb_000300_390 |
0.1165571971 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 6 |
Hb_152910_020 |
0.1211854985 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 7 |
Hb_002316_120 |
0.1247495606 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 8 |
Hb_007304_070 |
0.1261415477 |
- |
- |
hypoxanthine-guanine phosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_010897_030 |
0.1276441195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000896_120 |
0.1282476927 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 11 |
Hb_050806_010 |
0.1305021336 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 12 |
Hb_001017_070 |
0.13106385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005663_120 |
0.1325367592 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 14 |
Hb_183086_040 |
0.1351616209 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 15 |
Hb_000836_140 |
0.1367430802 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000692_020 |
0.1389784398 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 17 |
Hb_000389_080 |
0.1398419941 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 18 |
Hb_012539_020 |
0.1418536139 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 19 |
Hb_000820_130 |
0.1438882206 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_001377_380 |
0.1442823362 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |