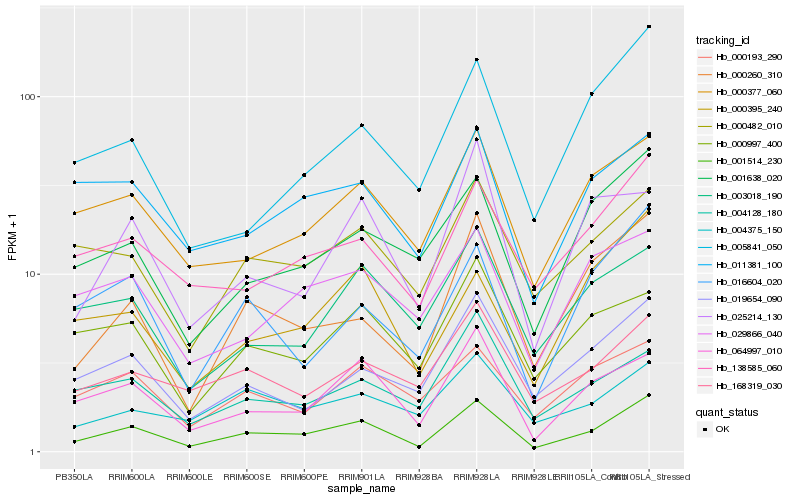
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_230 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 12-like [Jatropha curcas] |
| 2 |
Hb_064997_010 |
0.1325975621 |
- |
- |
hypothetical protein JCGZ_19852 [Jatropha curcas] |
| 3 |
Hb_000260_310 |
0.1421461866 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 4 |
Hb_000377_060 |
0.1523427321 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 5 |
Hb_138585_060 |
0.1525741646 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 6 |
Hb_001638_020 |
0.1552094138 |
- |
- |
PREDICTED: protein OSB1, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_016604_020 |
0.1553526304 |
- |
- |
- |
| 8 |
Hb_000482_010 |
0.1581767077 |
- |
- |
PREDICTED: probable ADP,ATP carrier protein At5g56450 [Jatropha curcas] |
| 9 |
Hb_019654_090 |
0.1584710749 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33680 [Jatropha curcas] |
| 10 |
Hb_000395_240 |
0.1589504909 |
- |
- |
hypothetical protein CICLE_v10010039mg [Citrus clementina] |
| 11 |
Hb_168319_030 |
0.1609688137 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_011381_100 |
0.1646524945 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2-like [Jatropha curcas] |
| 13 |
Hb_005841_050 |
0.1647597229 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 14 |
Hb_000193_290 |
0.1663896625 |
- |
- |
hypothetical protein JCGZ_11661 [Jatropha curcas] |
| 15 |
Hb_004375_150 |
0.1666458166 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 16 |
Hb_004128_180 |
0.1697793104 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_025214_130 |
0.1701642757 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 18 |
Hb_000997_400 |
0.1708779422 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g50270 [Jatropha curcas] |
| 19 |
Hb_029866_040 |
0.1711250062 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 20 |
Hb_003018_190 |
0.171478587 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g31070, mitochondrial [Jatropha curcas] |