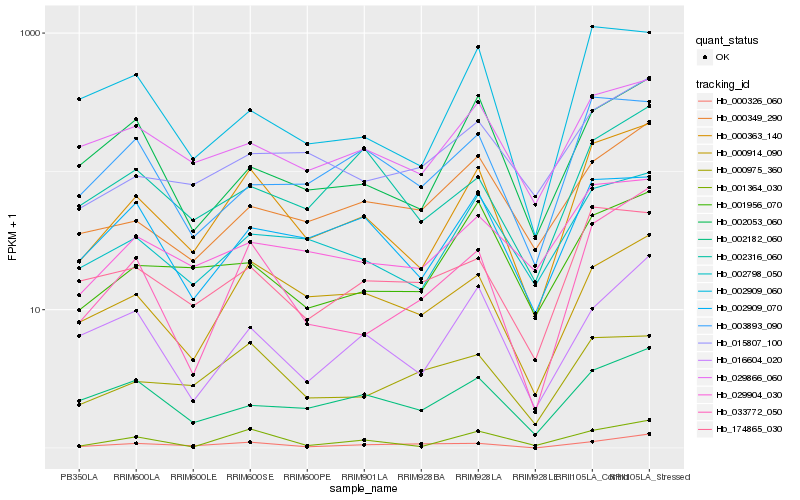
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_140 |
0.0 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 2 |
Hb_033772_050 |
0.1299816602 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 3 |
Hb_001364_030 |
0.1326975758 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 4 |
Hb_002798_050 |
0.1349687934 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 5 |
Hb_000914_090 |
0.1452480425 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 6 |
Hb_002909_070 |
0.1525443584 |
- |
- |
hypothetical protein POPTR_0006s03000g [Populus trichocarpa] |
| 7 |
Hb_174865_030 |
0.1544204609 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 8 |
Hb_029904_030 |
0.1544324719 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_029866_060 |
0.157626609 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 10 |
Hb_001956_070 |
0.1589845071 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002909_060 |
0.1622214011 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 12 |
Hb_016604_020 |
0.1636865151 |
- |
- |
- |
| 13 |
Hb_015807_100 |
0.1659758303 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000349_290 |
0.1660238874 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 15 |
Hb_002053_060 |
0.1671836649 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 16 |
Hb_003893_090 |
0.167725568 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 17 |
Hb_002182_060 |
0.167799658 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATXR6 [Jatropha curcas] |
| 18 |
Hb_000326_060 |
0.168480634 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 19 |
Hb_000975_360 |
0.1687526795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002316_060 |
0.1707057326 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |