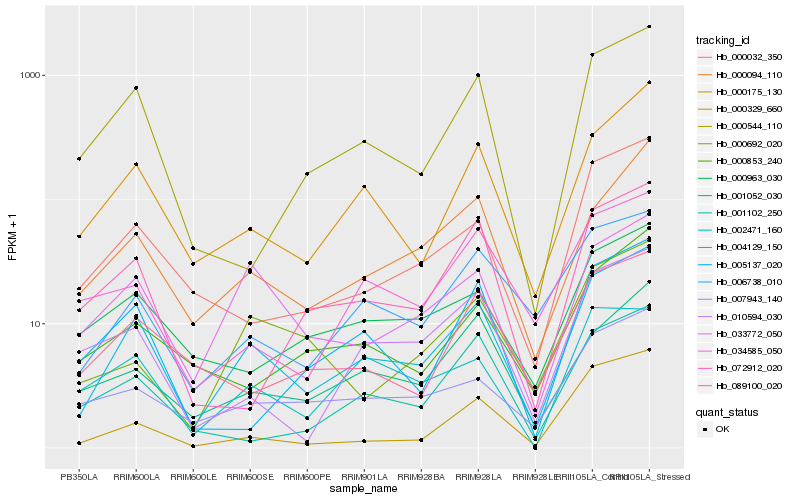
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_150 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 2 |
Hb_000175_130 |
0.1346559242 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 3 |
Hb_000094_110 |
0.1513780181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006738_010 |
0.1540273101 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 5 |
Hb_033772_050 |
0.1588713023 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 6 |
Hb_000032_350 |
0.1641990611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007943_140 |
0.1651305299 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |
| 8 |
Hb_000544_110 |
0.1684101101 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 9 |
Hb_001052_030 |
0.1719907796 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000329_660 |
0.1733533921 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 11 |
Hb_010594_030 |
0.1753273347 |
- |
- |
- |
| 12 |
Hb_089100_020 |
0.1789818161 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 13 |
Hb_000963_030 |
0.1801210526 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 14 |
Hb_000853_240 |
0.1805746216 |
- |
- |
- |
| 15 |
Hb_072912_020 |
0.181891539 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 16 |
Hb_005137_020 |
0.1820486336 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 17 |
Hb_001102_250 |
0.1836607385 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000692_020 |
0.1839718875 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 19 |
Hb_034585_050 |
0.1845728904 |
- |
- |
- |
| 20 |
Hb_002471_160 |
0.1860690929 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4-like [Populus euphratica] |