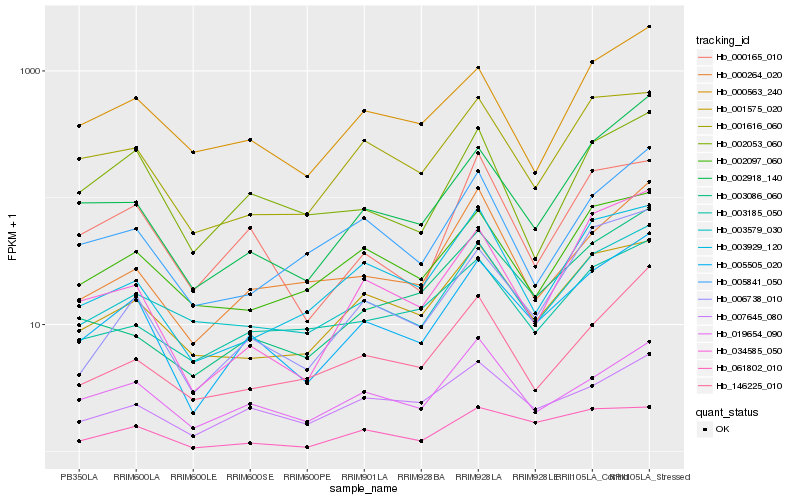
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005505_020 |
0.0 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |
| 2 |
Hb_002918_140 |
0.1239776259 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 3 |
Hb_006738_010 |
0.1277684874 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 4 |
Hb_000563_240 |
0.1303292636 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 5 |
Hb_003579_030 |
0.1307634719 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 6 |
Hb_007645_080 |
0.1309527587 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 7 |
Hb_034585_050 |
0.1324572537 |
- |
- |
- |
| 8 |
Hb_001575_020 |
0.1327895059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_019654_090 |
0.1342080712 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33680 [Jatropha curcas] |
| 10 |
Hb_000264_020 |
0.1384072769 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_146225_010 |
0.1389251784 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 12 |
Hb_005841_050 |
0.1415496467 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 13 |
Hb_003086_060 |
0.1415943042 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 14 |
Hb_001616_060 |
0.1425187451 |
- |
- |
PREDICTED: DNA-directed RNA polymerase subunit 10-like protein [Jatropha curcas] |
| 15 |
Hb_061802_010 |
0.1425487209 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 16 |
Hb_003929_120 |
0.1427414259 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000165_010 |
0.143691222 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 18 |
Hb_003185_050 |
0.1447985001 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 19 |
Hb_002097_060 |
0.1455062647 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002053_060 |
0.1478279 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |