| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
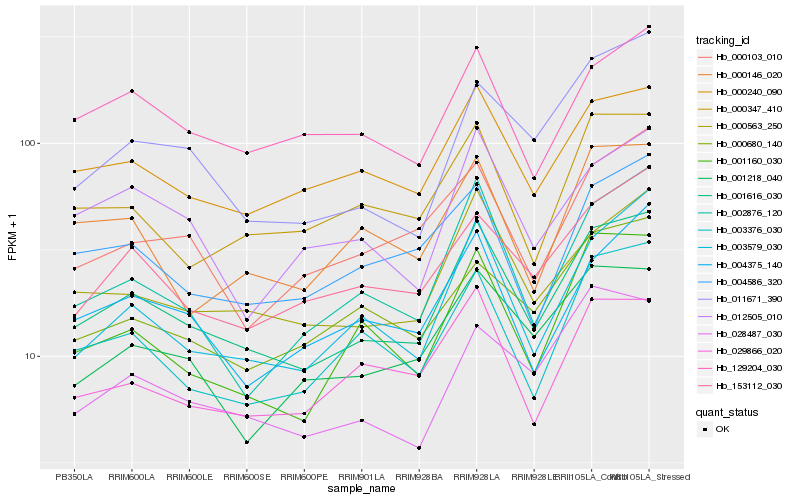
Hb_001616_030 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor-like protein At4g13040 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000563_250 |
0.071815771 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 3 |
Hb_000240_090 |
0.0789508493 |
- |
- |
Rab5 [Hevea brasiliensis] |
| 4 |
Hb_003579_030 |
0.084443326 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 5 |
Hb_003376_030 |
0.0865160242 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001160_030 |
0.0873492232 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein POPTR_0018s05470g [Populus trichocarpa] |
| 7 |
Hb_153112_030 |
0.0875152576 |
- |
- |
PREDICTED: UPF0678 fatty acid-binding protein-like protein At1g79260 [Jatropha curcas] |
| 8 |
Hb_002876_120 |
0.0912225829 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_028487_030 |
0.0919730601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001218_040 |
0.0922151614 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 11 |
Hb_004375_140 |
0.0936845756 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000347_410 |
0.0941527691 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 13 |
Hb_029866_020 |
0.0947114852 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |
| 14 |
Hb_004586_320 |
0.0950945064 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 15 |
Hb_011671_390 |
0.0966599398 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 16 |
Hb_129204_030 |
0.0975328355 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 17 |
Hb_000103_010 |
0.0975914032 |
- |
- |
20S proteasome beta subunit D2 [Hevea brasiliensis] |
| 18 |
Hb_000680_140 |
0.0984569557 |
- |
- |
PREDICTED: uncharacterized protein LOC105631472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012505_010 |
0.09857255 |
- |
- |
mak, putative [Ricinus communis] |
| 20 |
Hb_000146_020 |
0.0988754597 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |