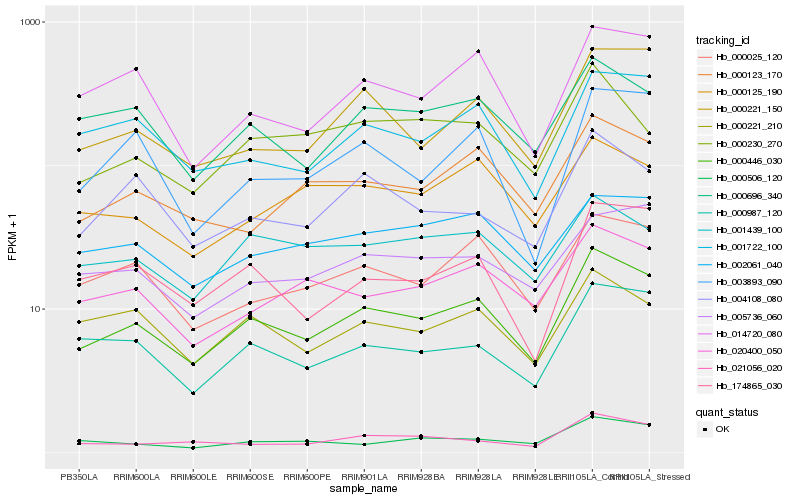
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000446_030 |
0.0 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000025_120 |
0.102549732 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000987_120 |
0.103333934 |
- |
- |
PREDICTED: protein SIEL [Jatropha curcas] |
| 4 |
Hb_001722_100 |
0.1074972964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000696_340 |
0.1076517009 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 6 |
Hb_174865_030 |
0.1092259613 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 7 |
Hb_000221_150 |
0.1096401663 |
- |
- |
- |
| 8 |
Hb_000123_170 |
0.1106623542 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 9 |
Hb_014720_080 |
0.1108225729 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 10 |
Hb_020400_050 |
0.1110851606 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 11 |
Hb_000221_210 |
0.1152715631 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_004108_080 |
0.1158802757 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase [Vitis vinifera] |
| 13 |
Hb_005736_060 |
0.1165942911 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 14 |
Hb_021056_020 |
0.1167837642 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 15 |
Hb_001439_100 |
0.1184577663 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 16 |
Hb_002061_040 |
0.1194410941 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 17 |
Hb_003893_090 |
0.1199516564 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 18 |
Hb_000125_190 |
0.1217039623 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000506_120 |
0.1224174993 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 20 |
Hb_000230_270 |
0.1226969579 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |