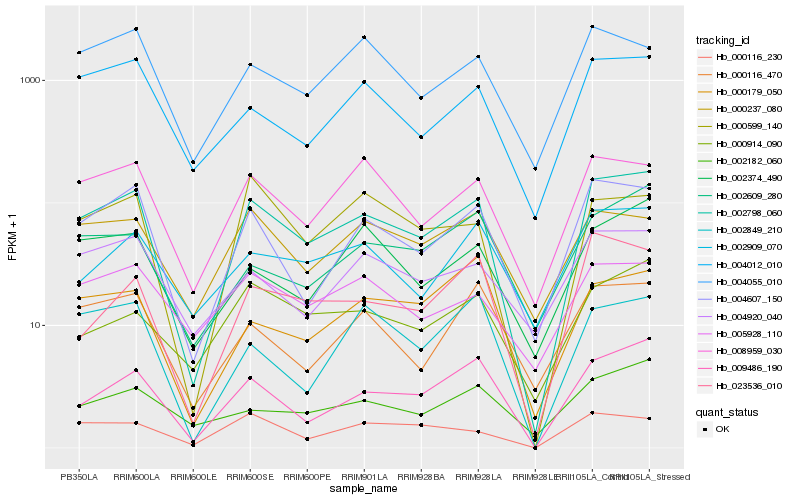
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002798_060 |
0.0 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 2 |
Hb_004607_150 |
0.1147715534 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 3 |
Hb_009486_190 |
0.1230844607 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_008959_030 |
0.1362043467 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 5 |
Hb_004012_010 |
0.1470687601 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1-like [Gossypium raimondii] |
| 6 |
Hb_023536_010 |
0.1491400268 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 7 |
Hb_000599_140 |
0.1502852816 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 4-like [Populus euphratica] |
| 8 |
Hb_000116_470 |
0.1543033312 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10-like [Jatropha curcas] |
| 9 |
Hb_000116_230 |
0.155993391 |
- |
- |
- |
| 10 |
Hb_004920_040 |
0.15623783 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 11 |
Hb_002374_490 |
0.1570630741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000237_080 |
0.1575295207 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 13 |
Hb_004055_010 |
0.1579408014 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_000914_090 |
0.1584614598 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 15 |
Hb_002909_070 |
0.1587030878 |
- |
- |
hypothetical protein POPTR_0006s03000g [Populus trichocarpa] |
| 16 |
Hb_002849_210 |
0.1607497686 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_002182_060 |
0.1607883093 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATXR6 [Jatropha curcas] |
| 18 |
Hb_005928_110 |
0.1610521098 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 19 |
Hb_002609_280 |
0.1615349278 |
- |
- |
villin 1-4, putative [Ricinus communis] |
| 20 |
Hb_000179_050 |
0.1632225607 |
- |
- |
40S ribosomal protein S17A [Hevea brasiliensis] |