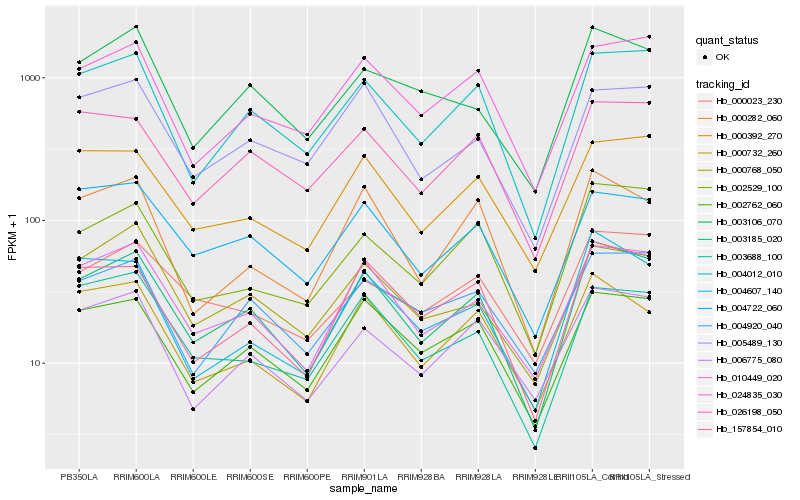
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003185_020 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 2 |
Hb_010449_020 |
0.0639444509 |
- |
- |
PREDICTED: rRNA-processing protein UTP23 homolog [Jatropha curcas] |
| 3 |
Hb_004012_010 |
0.0736929809 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1-like [Gossypium raimondii] |
| 4 |
Hb_157854_010 |
0.0811351764 |
- |
- |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 5 |
Hb_002762_060 |
0.0825903807 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 6 |
Hb_000392_270 |
0.0951018022 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 7 |
Hb_004722_060 |
0.0951109858 |
- |
- |
PREDICTED: protein CLP1 homolog [Jatropha curcas] |
| 8 |
Hb_026198_050 |
0.0956710819 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 9 |
Hb_006775_080 |
0.0964424043 |
- |
- |
PREDICTED: uncharacterized protein LOC105649730 [Jatropha curcas] |
| 10 |
Hb_024835_030 |
0.0969283496 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 11 |
Hb_002529_100 |
0.097225871 |
- |
- |
PREDICTED: DET1- and DDB1-associated protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000732_260 |
0.0982696589 |
- |
- |
PREDICTED: telomerase Cajal body protein 1 [Jatropha curcas] |
| 13 |
Hb_000282_060 |
0.0986937148 |
- |
- |
PREDICTED: uncharacterized protein LOC105638155 [Jatropha curcas] |
| 14 |
Hb_000768_050 |
0.0997201002 |
- |
- |
hypothetical protein RCOM_0845740 [Ricinus communis] |
| 15 |
Hb_003688_100 |
0.0999214624 |
- |
- |
Uncharacterized protein TCM_002515 [Theobroma cacao] |
| 16 |
Hb_005489_130 |
0.100138085 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 17 |
Hb_004920_040 |
0.1004735021 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 18 |
Hb_000023_230 |
0.1044937264 |
- |
- |
PREDICTED: uncharacterized protein LOC105637526 [Jatropha curcas] |
| 19 |
Hb_004607_140 |
0.1053098646 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 20 |
Hb_003106_070 |
0.1054160197 |
- |
- |
PREDICTED: uncharacterized protein LOC105167687 [Sesamum indicum] |