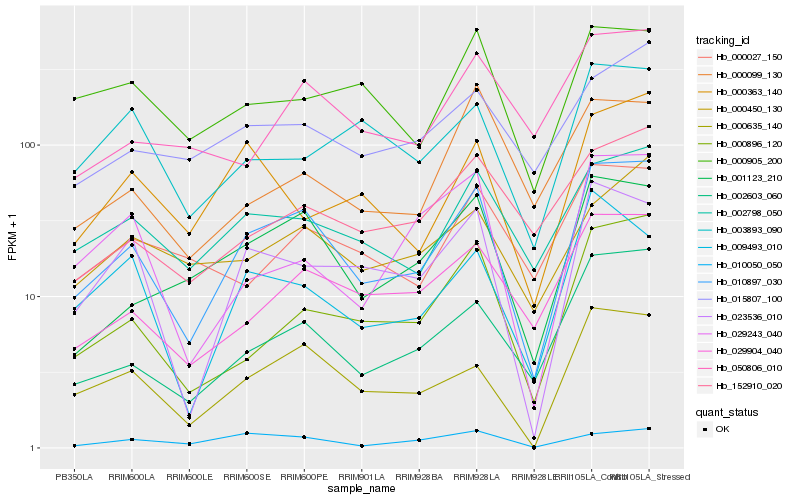
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010897_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000635_140 |
0.1234599932 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002603_060 |
0.1276441195 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 4 |
Hb_001123_210 |
0.1353033139 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 5 |
Hb_023536_010 |
0.1414516499 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 6 |
Hb_029904_040 |
0.1424063609 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 7 |
Hb_000896_120 |
0.1424839828 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 8 |
Hb_002798_050 |
0.1445836318 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 9 |
Hb_009493_010 |
0.1626354523 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 10 |
Hb_152910_020 |
0.1634032492 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 11 |
Hb_015807_100 |
0.1684829262 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_010050_050 |
0.1692318078 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_050806_010 |
0.17076535 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 14 |
Hb_000363_140 |
0.1711354205 |
- |
- |
hypothetical protein CICLE_v10009052mg [Citrus clementina] |
| 15 |
Hb_000027_150 |
0.1733101602 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 16 |
Hb_000905_200 |
0.1735555219 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 17 |
Hb_000099_130 |
0.1742972017 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 18 |
Hb_029243_040 |
0.1743792164 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Populus euphratica] |
| 19 |
Hb_000450_130 |
0.1767010973 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 20 |
Hb_003893_090 |
0.1775912732 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |