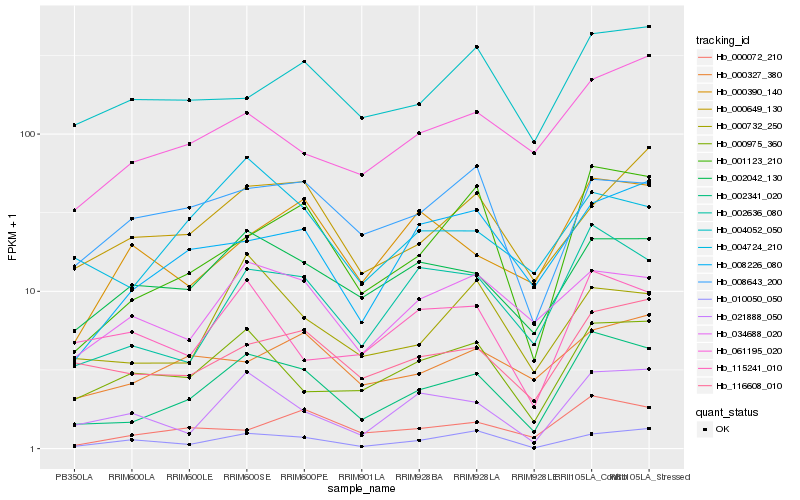
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002341_020 |
0.0 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 2 |
Hb_002636_080 |
0.1292831942 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 3 |
Hb_001123_210 |
0.1376539903 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 4 |
Hb_021888_050 |
0.1628058708 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 5 |
Hb_000975_360 |
0.1630039578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_116608_010 |
0.1634159832 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_002042_130 |
0.1652355993 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 8 |
Hb_000072_210 |
0.1687351599 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 9 |
Hb_008226_080 |
0.1717537385 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 10 |
Hb_000732_250 |
0.1781462286 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 11 |
Hb_010050_050 |
0.1791014791 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_034688_020 |
0.1815831105 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 13 |
Hb_061195_020 |
0.1826995492 |
- |
- |
PREDICTED: uncharacterized protein LOC101990594 [Microtus ochrogaster] |
| 14 |
Hb_004724_210 |
0.1843742424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_115241_010 |
0.1862826509 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 16 |
Hb_004052_050 |
0.1879057737 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |
| 17 |
Hb_000390_140 |
0.1895081176 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 18 |
Hb_000327_380 |
0.1913164444 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 19 |
Hb_008643_200 |
0.1934358112 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 [Vitis vinifera] |
| 20 |
Hb_000649_130 |
0.1938741899 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |