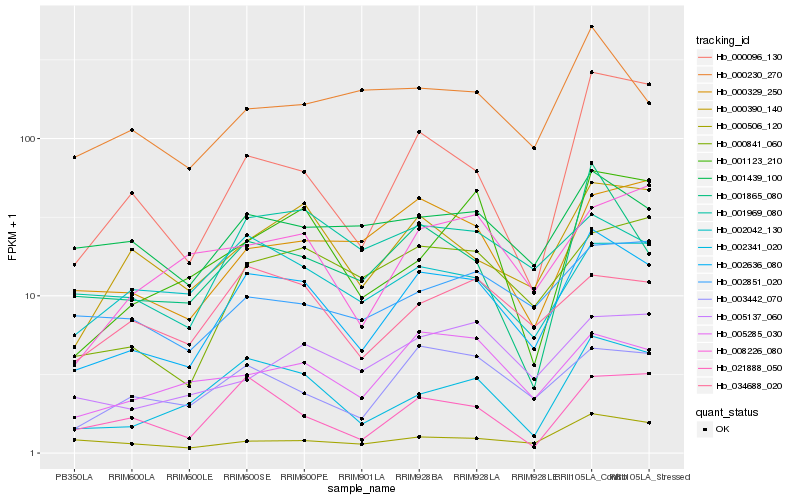
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002636_080 |
0.0 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 2 |
Hb_002341_020 |
0.1292831942 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 3 |
Hb_003442_070 |
0.1426148446 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 4 |
Hb_000390_140 |
0.1440428935 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 5 |
Hb_000841_060 |
0.1487820109 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0015s06470g [Populus trichocarpa] |
| 6 |
Hb_005285_030 |
0.1515722758 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 7 |
Hb_034688_020 |
0.1575879126 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 8 |
Hb_005137_060 |
0.165011416 |
- |
- |
- |
| 9 |
Hb_001439_100 |
0.1650606141 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 10 |
Hb_001865_080 |
0.1652089255 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 11 |
Hb_000329_250 |
0.1673862767 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_021888_050 |
0.1687591484 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 13 |
Hb_002042_130 |
0.16971879 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 14 |
Hb_000230_270 |
0.170604963 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 15 |
Hb_002851_020 |
0.1708345673 |
- |
- |
PREDICTED: uncharacterized protein LOC105633802 [Jatropha curcas] |
| 16 |
Hb_000096_130 |
0.170959143 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 17 |
Hb_008226_080 |
0.1720356202 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 18 |
Hb_001969_080 |
0.1722595128 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 19 |
Hb_000506_120 |
0.1731481651 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 20 |
Hb_001123_210 |
0.1739465655 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |