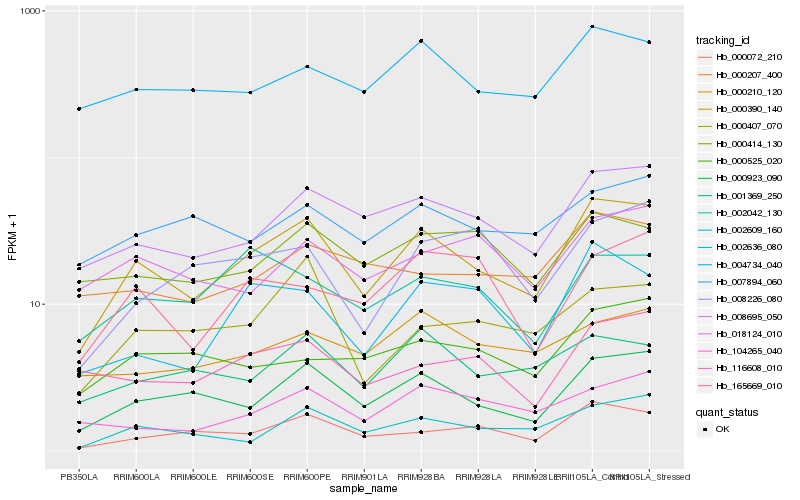
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_140 |
0.0 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 2 |
Hb_000923_090 |
0.1061080732 |
- |
- |
- |
| 3 |
Hb_008695_050 |
0.1204367481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000072_210 |
0.1350349736 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 5 |
Hb_002609_160 |
0.1435037102 |
- |
- |
PREDICTED: GDP-mannose transporter GONST2 [Jatropha curcas] |
| 6 |
Hb_002636_080 |
0.1440428935 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 7 |
Hb_004734_040 |
0.1457407125 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |
| 8 |
Hb_001369_250 |
0.1538841449 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 9 |
Hb_104265_040 |
0.1572441126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_116608_010 |
0.1578696918 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000207_400 |
0.1587755371 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000414_130 |
0.1588534848 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 13 |
Hb_000407_070 |
0.1609725919 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007894_060 |
0.1610494356 |
- |
- |
PREDICTED: ubiquitin thioesterase OTU1 [Jatropha curcas] |
| 15 |
Hb_018124_010 |
0.1620541 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002042_130 |
0.1621671817 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 17 |
Hb_000525_020 |
0.1623482 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_165669_010 |
0.1626218855 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_008226_080 |
0.1646853416 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 20 |
Hb_000210_120 |
0.164691094 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |