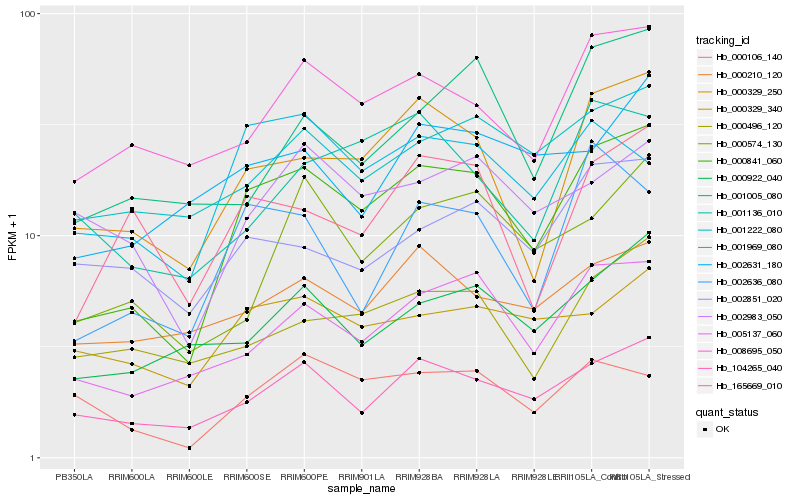
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000841_060 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0015s06470g [Populus trichocarpa] |
| 2 |
Hb_000329_250 |
0.1225560415 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_104265_040 |
0.1243124728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005137_060 |
0.124669156 |
- |
- |
- |
| 5 |
Hb_008695_050 |
0.1431354743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002636_080 |
0.1487820109 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 7 |
Hb_001222_080 |
0.1495271933 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 8 |
Hb_165669_010 |
0.1509518552 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000922_040 |
0.1513550021 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 10 |
Hb_001969_080 |
0.1572656515 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 11 |
Hb_000329_340 |
0.1582681454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000574_130 |
0.1586782419 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 13 |
Hb_000496_120 |
0.1598322234 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 14 |
Hb_000106_140 |
0.1610327552 |
- |
- |
- |
| 15 |
Hb_000210_120 |
0.1611768029 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002983_050 |
0.1635980099 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001136_010 |
0.1645818452 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 18 |
Hb_002851_020 |
0.1650971779 |
- |
- |
PREDICTED: uncharacterized protein LOC105633802 [Jatropha curcas] |
| 19 |
Hb_002631_180 |
0.1658359304 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 20 |
Hb_001005_080 |
0.1663720742 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |