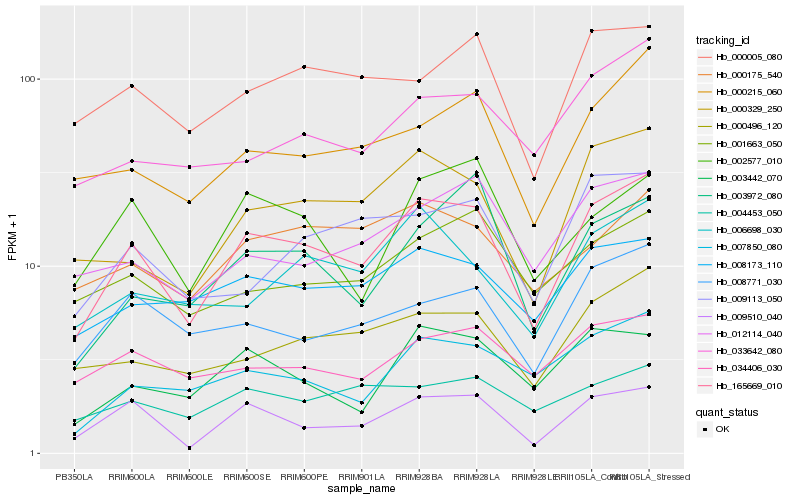
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165669_010 |
0.0 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_009510_040 |
0.1107068957 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000329_250 |
0.1144786431 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000496_120 |
0.1233283941 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 5 |
Hb_000215_060 |
0.1268716673 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 6 |
Hb_008173_110 |
0.128892135 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_008771_030 |
0.1290030563 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007850_080 |
0.1315332484 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012114_040 |
0.1334660765 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_003442_070 |
0.1340904061 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 11 |
Hb_000005_080 |
0.1349547626 |
- |
- |
PREDICTED: cyclin-dependent kinases regulatory subunit 1 [Elaeis guineensis] |
| 12 |
Hb_006698_030 |
0.1360433672 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 13 |
Hb_033642_080 |
0.1378760324 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 14 |
Hb_034406_030 |
0.1383920694 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_009113_050 |
0.1387940178 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 16 |
Hb_002577_010 |
0.1397394446 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 17 |
Hb_001663_050 |
0.1399035568 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 18 |
Hb_000175_540 |
0.1400716938 |
- |
- |
PREDICTED: uncharacterized protein LOC105634886 [Jatropha curcas] |
| 19 |
Hb_004453_050 |
0.1404969988 |
- |
- |
PREDICTED: uncharacterized protein LOC105637675 [Jatropha curcas] |
| 20 |
Hb_003972_080 |
0.1424916774 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |