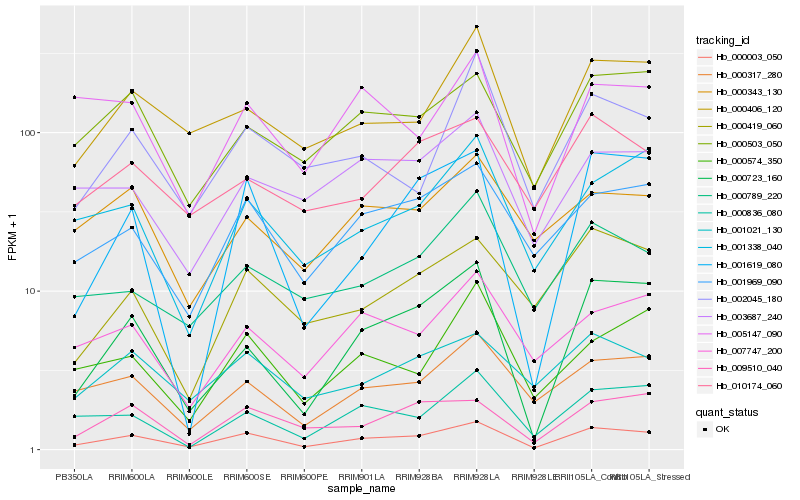
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_050 |
0.0 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 2 |
Hb_000723_160 |
0.1239588276 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 3 |
Hb_001969_090 |
0.1323226942 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 4 |
Hb_000317_280 |
0.141454886 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At5g08490 [Jatropha curcas] |
| 5 |
Hb_000406_120 |
0.1448698462 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_009510_040 |
0.1462213477 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000503_050 |
0.1466730332 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 1-like [Gossypium raimondii] |
| 8 |
Hb_000836_080 |
0.1481314582 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g46050, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001619_080 |
0.1485690328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001338_040 |
0.1528034267 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_007747_200 |
0.1546677302 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g37570 [Jatropha curcas] |
| 12 |
Hb_001021_130 |
0.1549642848 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 13 |
Hb_000574_350 |
0.1570272742 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 14 |
Hb_000419_060 |
0.158458278 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 15 |
Hb_000789_220 |
0.160887929 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 16 |
Hb_002045_180 |
0.1629948548 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 17 |
Hb_003687_240 |
0.1633614383 |
- |
- |
PREDICTED: cryptochrome-1 [Jatropha curcas] |
| 18 |
Hb_000343_130 |
0.1667968821 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 19 |
Hb_005147_090 |
0.1683761024 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010174_060 |
0.1685871592 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |