| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
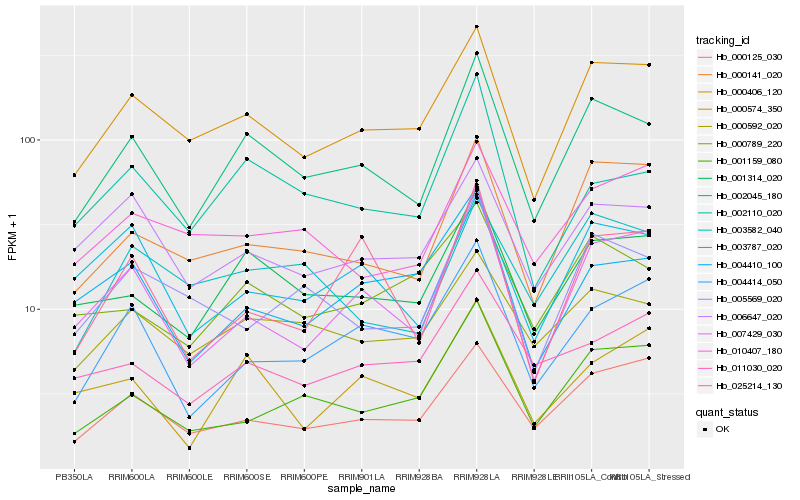
Hb_002045_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 2 |
Hb_001314_020 |
0.0979982596 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000406_120 |
0.1036430588 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000592_020 |
0.1187878064 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 5 |
Hb_000141_020 |
0.124532148 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 6 |
Hb_007429_030 |
0.1281261771 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002110_020 |
0.1312923693 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 8 |
Hb_000125_030 |
0.1328396911 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 9 |
Hb_000789_220 |
0.1353759749 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 10 |
Hb_006647_020 |
0.1374519698 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 11 |
Hb_003787_020 |
0.1387569403 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit A [Jatropha curcas] |
| 12 |
Hb_003582_040 |
0.1391879077 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 13 |
Hb_005569_020 |
0.1413215807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001159_080 |
0.1425278799 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 15 |
Hb_004414_050 |
0.1458676015 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000574_350 |
0.14854722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 17 |
Hb_011030_020 |
0.1528193365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 18 |
Hb_010407_180 |
0.1530953547 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 19 |
Hb_025214_130 |
0.1538194171 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 20 |
Hb_004410_100 |
0.1541999648 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |