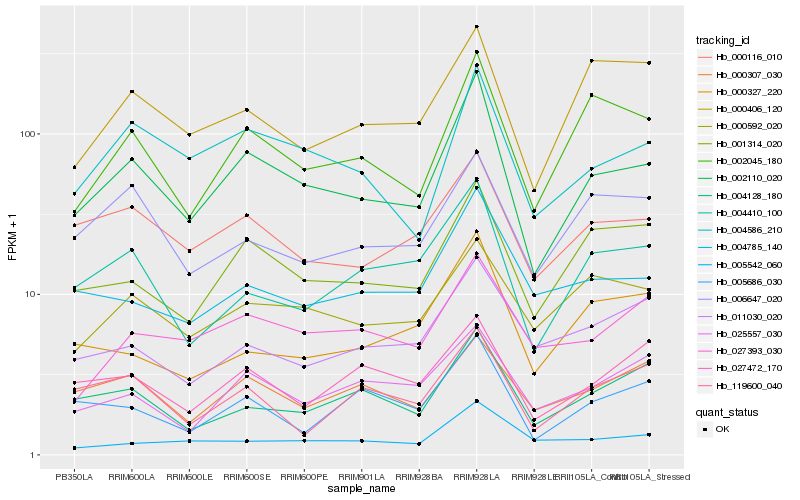
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_020 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 2 |
Hb_004586_210 |
0.1169938542 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002045_180 |
0.1312923693 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 4 |
Hb_001314_020 |
0.1361338853 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000116_010 |
0.1382914809 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_004410_100 |
0.1446000753 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 7 |
Hb_004785_140 |
0.1454740541 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_004128_180 |
0.1525715397 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000592_020 |
0.1566305544 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 10 |
Hb_005542_060 |
0.15849199 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005686_030 |
0.1586862255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_027393_030 |
0.1589621586 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |
| 13 |
Hb_000307_030 |
0.1591656078 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g40410, mitochondrial [Jatropha curcas] |
| 14 |
Hb_025557_030 |
0.1614065582 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_027472_170 |
0.1628066527 |
- |
- |
hypothetical protein POPTR_0007s08080g [Populus trichocarpa] |
| 16 |
Hb_000406_120 |
0.1630172198 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_006647_020 |
0.1643285824 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 18 |
Hb_119600_040 |
0.1670922483 |
- |
- |
hypothetical protein JCGZ_08045 [Jatropha curcas] |
| 19 |
Hb_000327_220 |
0.1684243176 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 20 |
Hb_011030_020 |
0.1685441379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |