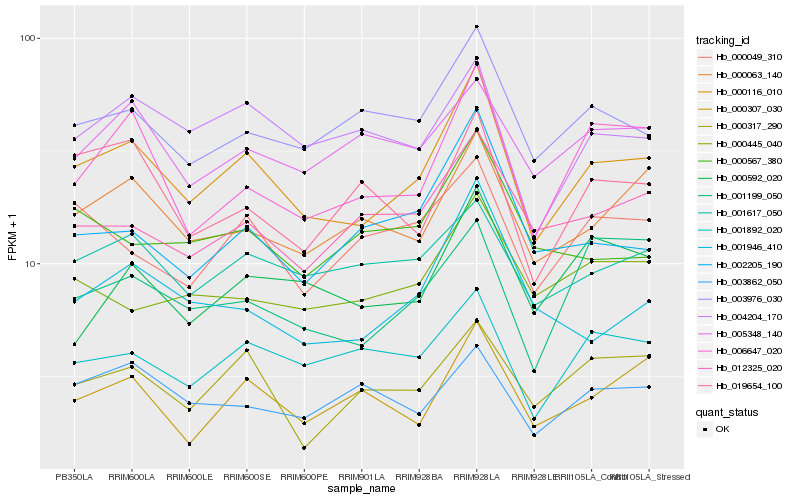
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_010 |
0.0 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_006647_020 |
0.1075471341 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 3 |
Hb_003976_030 |
0.1090647175 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase beta subunit [Jatropha curcas] |
| 4 |
Hb_000063_140 |
0.1095608102 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 25 kDa protein [Jatropha curcas] |
| 5 |
Hb_001617_050 |
0.1143276555 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001892_020 |
0.1150719083 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_004204_170 |
0.1172535734 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 8 |
Hb_012325_020 |
0.1196398435 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001946_410 |
0.12026654 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000049_310 |
0.1206542755 |
- |
- |
Myosin-J heavy chain, partial [Glycine soja] |
| 11 |
Hb_000317_290 |
0.121633281 |
- |
- |
PREDICTED: VQ motif-containing protein 31 [Jatropha curcas] |
| 12 |
Hb_002205_190 |
0.1220263793 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 13 |
Hb_000445_040 |
0.1247753128 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000307_030 |
0.1269196137 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g40410, mitochondrial [Jatropha curcas] |
| 15 |
Hb_019654_100 |
0.1278964249 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001199_050 |
0.1288209293 |
- |
- |
PREDICTED: DNA-3-methyladenine glycosylase-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000592_020 |
0.1294962627 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 18 |
Hb_005348_140 |
0.1297778719 |
- |
- |
PREDICTED: ubiquinone biosynthesis protein COQ9-B, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000567_380 |
0.1313536201 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 20 |
Hb_003862_050 |
0.1327588567 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |