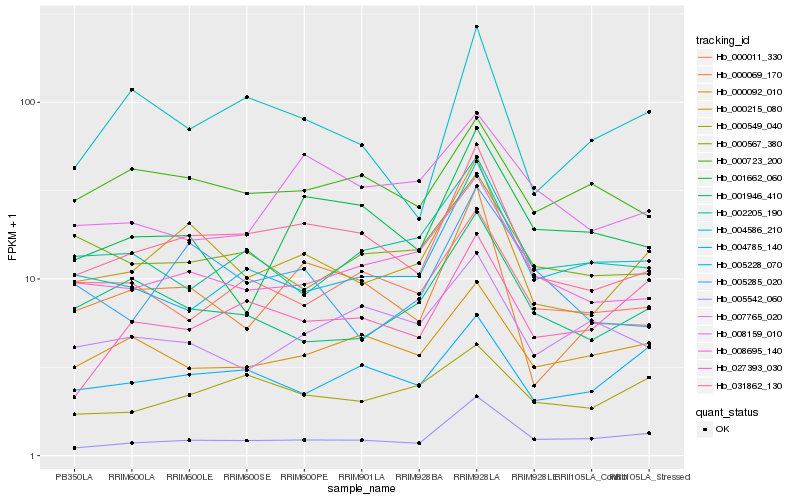
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_130 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000215_080 |
0.1192219876 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000069_170 |
0.1226010229 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 4 |
Hb_008695_140 |
0.1263771027 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 5 |
Hb_000011_330 |
0.1337780783 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 6 |
Hb_000723_200 |
0.138080066 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005542_060 |
0.1441384876 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000092_010 |
0.1463152414 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 9 |
Hb_004586_210 |
0.1491777541 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001662_060 |
0.1517903539 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000567_380 |
0.1547081931 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 12 |
Hb_007765_020 |
0.1594907966 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 13 |
Hb_005228_070 |
0.1597750946 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g13230, mitochondrial [Jatropha curcas] |
| 14 |
Hb_004785_140 |
0.1598888345 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_005285_020 |
0.1600804039 |
- |
- |
- |
| 16 |
Hb_002205_190 |
0.1606917209 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 17 |
Hb_008159_010 |
0.1612792441 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 18 |
Hb_027393_030 |
0.1638501092 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |
| 19 |
Hb_000549_040 |
0.1644270825 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001946_410 |
0.1664823012 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |