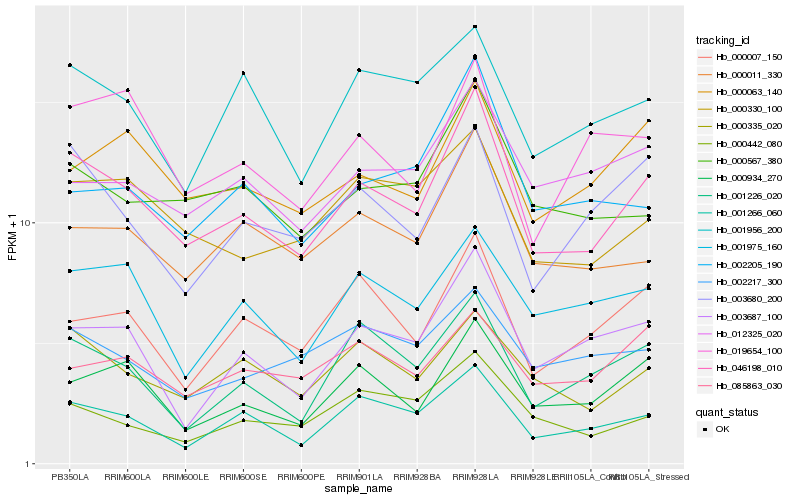
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046198_010 |
0.0 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 2 |
Hb_000335_020 |
0.1021779244 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000011_330 |
0.1037452527 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 4 |
Hb_001266_060 |
0.1042172147 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 5 |
Hb_000567_380 |
0.1068096725 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 6 |
Hb_000330_100 |
0.1118273295 |
- |
- |
hypothetical protein VITISV_011650 [Vitis vinifera] |
| 7 |
Hb_000063_140 |
0.1131933777 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 25 kDa protein [Jatropha curcas] |
| 8 |
Hb_000934_270 |
0.1132081934 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g21470 [Populus euphratica] |
| 9 |
Hb_003680_200 |
0.1135765562 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 10 |
Hb_000007_150 |
0.1171380014 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41080 [Jatropha curcas] |
| 11 |
Hb_001975_160 |
0.1182960724 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_012325_020 |
0.1202908246 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003687_100 |
0.1205299748 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 14 |
Hb_001956_200 |
0.1215533971 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_001226_020 |
0.1217034605 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_002217_300 |
0.124251532 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 17 |
Hb_019654_100 |
0.1244115539 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_002205_190 |
0.1249446594 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 19 |
Hb_000442_080 |
0.1258887482 |
- |
- |
hypothetical protein JCGZ_03373 [Jatropha curcas] |
| 20 |
Hb_085863_030 |
0.1261845836 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g31430 [Jatropha curcas] |