| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
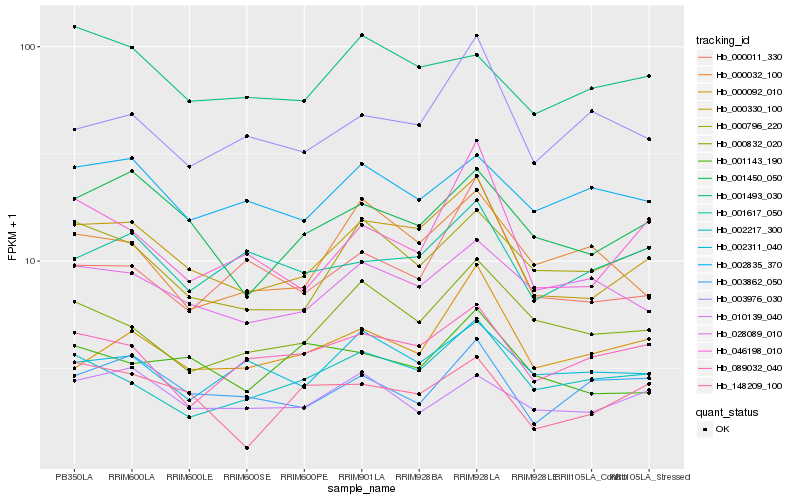
Hb_000330_100 |
0.0 |
- |
- |
hypothetical protein VITISV_011650 [Vitis vinifera] |
| 2 |
Hb_001450_050 |
0.0971156844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000796_220 |
0.1082626525 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 4 |
Hb_002217_300 |
0.1096110234 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 5 |
Hb_046198_010 |
0.1118273295 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 6 |
Hb_000832_020 |
0.1131873031 |
transcription factor |
TF Family: B3 |
hypothetical protein EUGRSUZ_D00268 [Eucalyptus grandis] |
| 7 |
Hb_028089_010 |
0.114902131 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |
| 8 |
Hb_089032_040 |
0.1177533483 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80550, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001617_050 |
0.1179957333 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000092_010 |
0.1209996761 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 11 |
Hb_002311_040 |
0.1210136054 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 12 |
Hb_000011_330 |
0.1211239569 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 13 |
Hb_003976_030 |
0.1222220263 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase beta subunit [Jatropha curcas] |
| 14 |
Hb_148209_100 |
0.1222810632 |
- |
- |
phosphatidylinositol 4-kinase, putative [Ricinus communis] |
| 15 |
Hb_001143_190 |
0.1225299983 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 16 |
Hb_010139_040 |
0.1229017558 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001493_030 |
0.122914902 |
- |
- |
hypothetical protein MIMGU_mgv1a005351mg [Erythranthe guttata] |
| 18 |
Hb_003862_050 |
0.122932759 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 19 |
Hb_000032_100 |
0.1230199241 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Vitis vinifera] |
| 20 |
Hb_002835_370 |
0.1233144708 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |