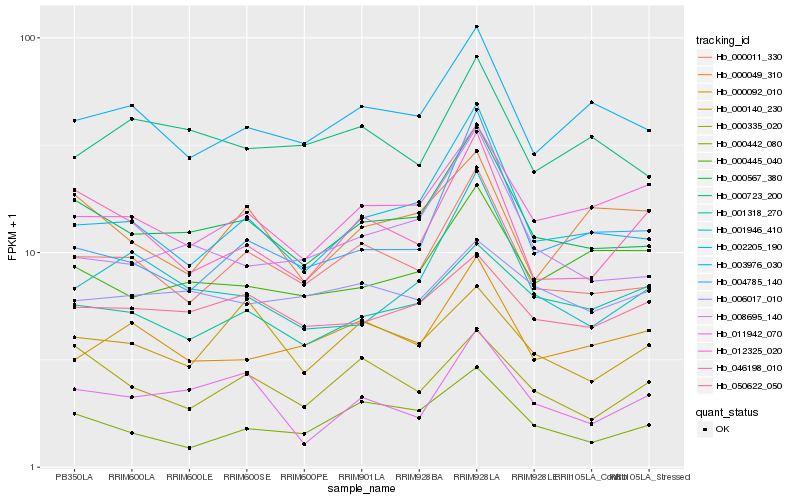
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_380 |
0.0 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 2 |
Hb_000011_330 |
0.0716522956 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 3 |
Hb_002205_190 |
0.0797470296 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 4 |
Hb_008695_140 |
0.0934444783 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 5 |
Hb_003976_030 |
0.0964896602 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase beta subunit [Jatropha curcas] |
| 6 |
Hb_012325_020 |
0.1017575524 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_050622_050 |
0.105814706 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 8 |
Hb_046198_010 |
0.1068096725 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 9 |
Hb_000445_040 |
0.1084401762 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001318_270 |
0.115059193 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_001946_410 |
0.1158353425 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000723_200 |
0.1189360469 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000335_020 |
0.1224476322 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000049_310 |
0.122587396 |
- |
- |
Myosin-J heavy chain, partial [Glycine soja] |
| 15 |
Hb_011942_070 |
0.1227215958 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 16 |
Hb_000092_010 |
0.1233390196 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 17 |
Hb_004785_140 |
0.1237072029 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000442_080 |
0.1249492222 |
- |
- |
hypothetical protein JCGZ_03373 [Jatropha curcas] |
| 19 |
Hb_006017_010 |
0.1250118039 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 20 |
Hb_000140_230 |
0.1251218104 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |