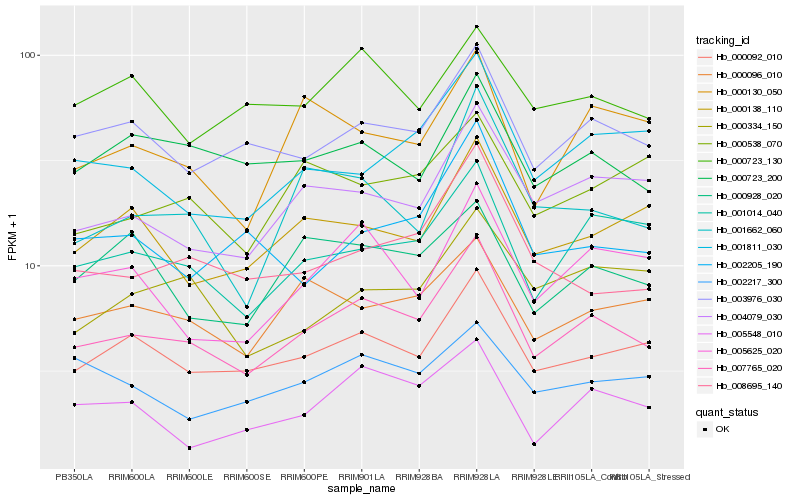
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 2 |
Hb_000092_010 |
0.0882879873 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 3 |
Hb_003976_030 |
0.0957400208 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase beta subunit [Jatropha curcas] |
| 4 |
Hb_001014_040 |
0.1007747496 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 5 |
Hb_004079_030 |
0.1010863078 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001662_060 |
0.1112192551 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_005625_020 |
0.111457093 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008695_140 |
0.1124340456 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 9 |
Hb_000096_010 |
0.1127507582 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 10 |
Hb_000723_200 |
0.1135578991 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001811_030 |
0.118333804 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 12 |
Hb_000138_110 |
0.1189801061 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 13 |
Hb_000130_050 |
0.1189872529 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 14 |
Hb_000723_130 |
0.124400226 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 15 |
Hb_005548_010 |
0.1246022189 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 16 |
Hb_002205_190 |
0.1265834103 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 17 |
Hb_002217_300 |
0.1266792822 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 18 |
Hb_000928_020 |
0.1271916051 |
- |
- |
hypothetical protein CICLE_v10013682mg [Citrus clementina] |
| 19 |
Hb_000334_150 |
0.1275872966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000538_070 |
0.1288929092 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |