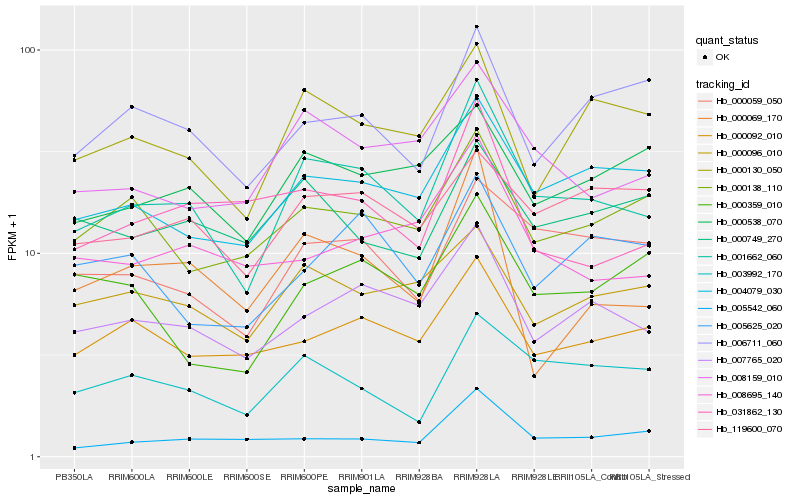
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_060 |
0.0 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_007765_020 |
0.1112192551 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 3 |
Hb_004079_030 |
0.1202915006 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008159_010 |
0.1233832565 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 5 |
Hb_000059_050 |
0.1306479351 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 6 |
Hb_000130_050 |
0.1379669448 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 7 |
Hb_000092_010 |
0.1403229222 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 8 |
Hb_003992_170 |
0.1403893345 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 9 |
Hb_000096_010 |
0.1440528378 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 10 |
Hb_000138_110 |
0.1448004273 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 11 |
Hb_008695_140 |
0.1456592389 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 12 |
Hb_000538_070 |
0.1506219173 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 13 |
Hb_119600_070 |
0.1514723903 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 14 |
Hb_031862_130 |
0.1517903539 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_006711_060 |
0.1532995515 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 16 |
Hb_000069_170 |
0.1533548743 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 17 |
Hb_005542_060 |
0.1548258892 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_005625_020 |
0.1581543823 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000749_270 |
0.1603187361 |
- |
- |
PREDICTED: WD repeat-containing protein 44 [Jatropha curcas] |
| 20 |
Hb_000359_010 |
0.1607960569 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |