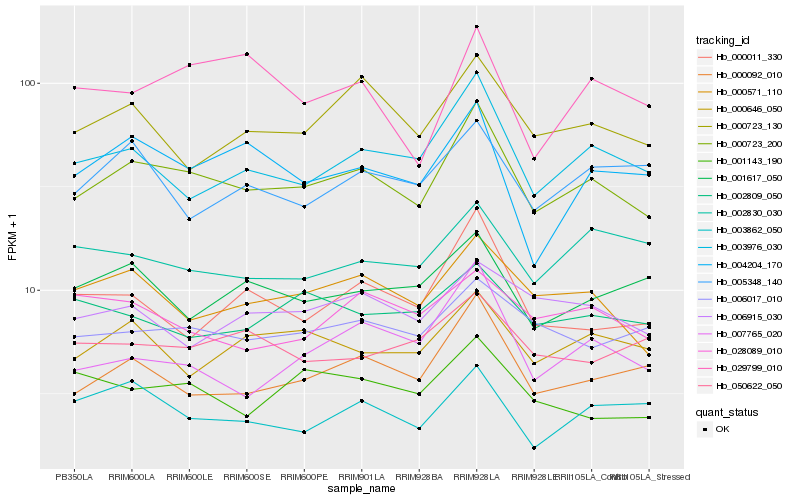
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_200 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003976_030 |
0.0879369818 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase beta subunit [Jatropha curcas] |
| 3 |
Hb_000571_110 |
0.0898999501 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |
| 4 |
Hb_000092_010 |
0.0934732166 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646502 [Jatropha curcas] |
| 5 |
Hb_000723_130 |
0.1000411091 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 6 |
Hb_006017_010 |
0.1020445252 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 7 |
Hb_004204_170 |
0.1025353611 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 8 |
Hb_000646_050 |
0.1038455659 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 9 |
Hb_000011_330 |
0.1094047538 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 10 |
Hb_006915_030 |
0.1119751656 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |
| 11 |
Hb_007765_020 |
0.1135578991 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 12 |
Hb_003862_050 |
0.1137171987 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 13 |
Hb_001617_050 |
0.1137758825 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005348_140 |
0.1147434719 |
- |
- |
PREDICTED: ubiquinone biosynthesis protein COQ9-B, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001143_190 |
0.115242703 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 16 |
Hb_029799_010 |
0.1162625034 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_002830_030 |
0.1168164983 |
- |
- |
PREDICTED: CDK5RAP3-like protein [Jatropha curcas] |
| 18 |
Hb_050622_050 |
0.1177964419 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 19 |
Hb_002809_050 |
0.1180825145 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 20 |
Hb_028089_010 |
0.1182365077 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |