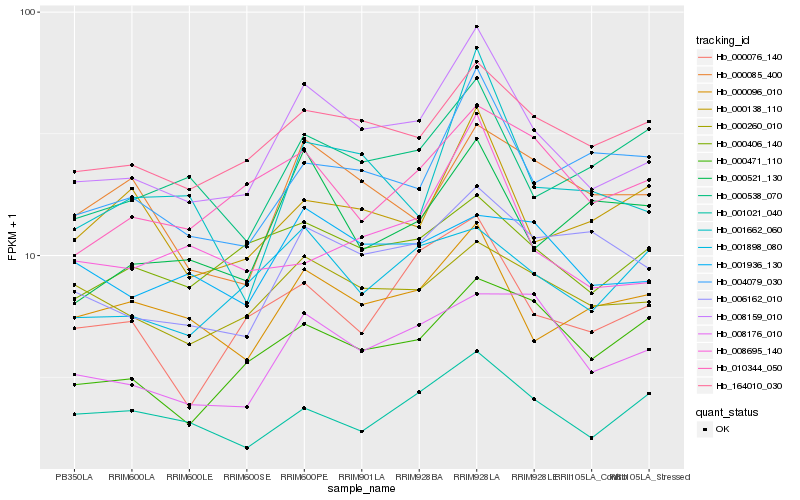
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008159_010 |
0.0 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 2 |
Hb_164010_030 |
0.1121111853 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 3 |
Hb_001662_060 |
0.1233832565 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_000096_010 |
0.1242171591 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 5 |
Hb_004079_030 |
0.1246620511 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006162_010 |
0.1251673105 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |
| 7 |
Hb_000076_140 |
0.1256194488 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_008176_010 |
0.1282333901 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 9 |
Hb_010344_050 |
0.1303604642 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 10 |
Hb_000260_010 |
0.1304469954 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 11 |
Hb_000406_140 |
0.1313394159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000538_070 |
0.1318044784 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 13 |
Hb_008695_140 |
0.1335795122 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 14 |
Hb_000138_110 |
0.1371317818 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 15 |
Hb_000471_110 |
0.1380165882 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000521_130 |
0.1382361659 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001898_080 |
0.1389200508 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000085_400 |
0.1392622629 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 19 |
Hb_001936_130 |
0.140000248 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 20 |
Hb_001021_040 |
0.141073227 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |