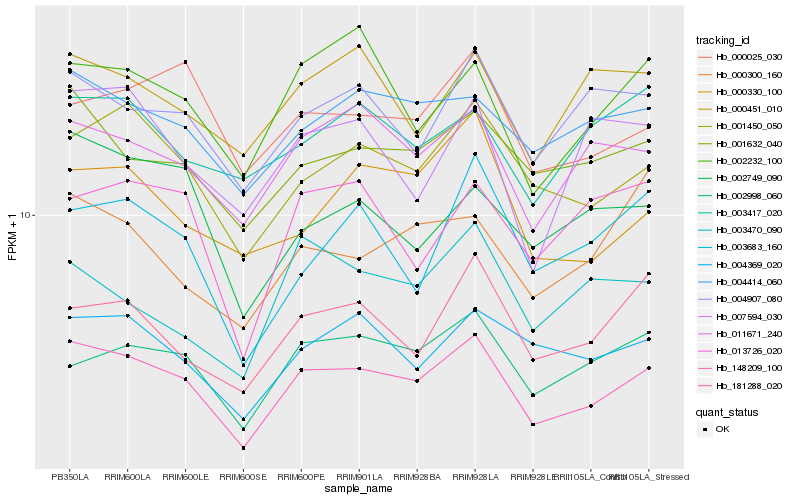
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148209_100 |
0.0 |
- |
- |
phosphatidylinositol 4-kinase, putative [Ricinus communis] |
| 2 |
Hb_001450_050 |
0.0973973553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002998_060 |
0.1055565988 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 4 |
Hb_011671_240 |
0.1085557862 |
- |
- |
PREDICTED: WD repeat-containing protein WRAP73-like [Jatropha curcas] |
| 5 |
Hb_001632_040 |
0.113106827 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_002232_100 |
0.1136379729 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 7 |
Hb_004369_020 |
0.1138815656 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 8 |
Hb_004414_060 |
0.1148806252 |
- |
- |
hypothetical protein JCGZ_17225 [Jatropha curcas] |
| 9 |
Hb_181288_020 |
0.1162867099 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 10 |
Hb_004907_080 |
0.1165102523 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_013726_020 |
0.1216121778 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_003683_160 |
0.1218251473 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 13 |
Hb_000330_100 |
0.1222810632 |
- |
- |
hypothetical protein VITISV_011650 [Vitis vinifera] |
| 14 |
Hb_000025_030 |
0.1227210734 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_007594_030 |
0.1240042023 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 16 |
Hb_002749_090 |
0.1249872882 |
- |
- |
- |
| 17 |
Hb_000451_010 |
0.127649135 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 18 |
Hb_003417_020 |
0.130370347 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003470_090 |
0.1317325948 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 20 |
Hb_000300_160 |
0.1322754657 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |