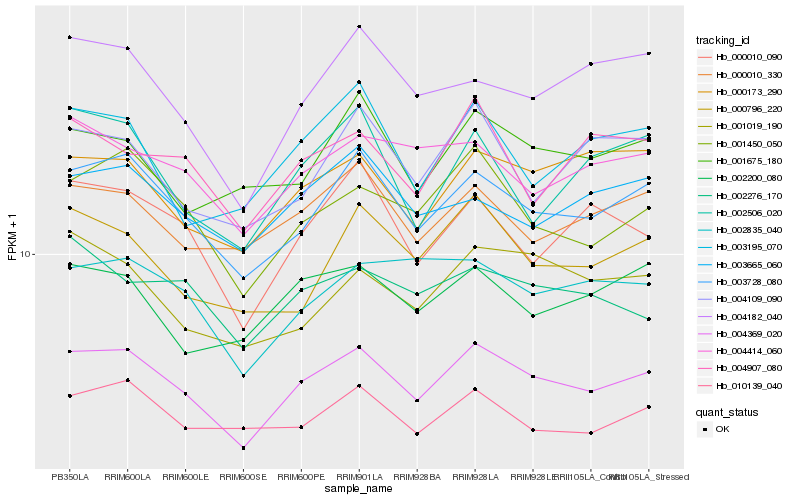
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004369_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 2 |
Hb_003728_080 |
0.0556251282 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like [Jatropha curcas] |
| 3 |
Hb_001450_050 |
0.0757484988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000010_090 |
0.0885199487 |
- |
- |
PREDICTED: afadin- and alpha-actinin-binding protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_000173_290 |
0.0894411532 |
- |
- |
guanylate kinase, putative [Ricinus communis] |
| 6 |
Hb_004182_040 |
0.0899559158 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Jatropha curcas] |
| 7 |
Hb_000010_330 |
0.0914869948 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 8 |
Hb_000796_220 |
0.0935834268 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 9 |
Hb_010139_040 |
0.0936773638 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_004414_060 |
0.0948336543 |
- |
- |
hypothetical protein JCGZ_17225 [Jatropha curcas] |
| 11 |
Hb_002835_040 |
0.0956041113 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001019_190 |
0.0956867426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002200_080 |
0.0964007842 |
- |
- |
PREDICTED: uncharacterized protein C57A10.07 [Jatropha curcas] |
| 14 |
Hb_003665_060 |
0.0977523228 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 15 |
Hb_003195_070 |
0.0980909972 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_002506_020 |
0.0989884042 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_001675_180 |
0.0991726167 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 18 |
Hb_004109_090 |
0.0997471872 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_002276_170 |
0.1004771136 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004907_080 |
0.1005866697 |
- |
- |
zinc finger protein, putative [Ricinus communis] |