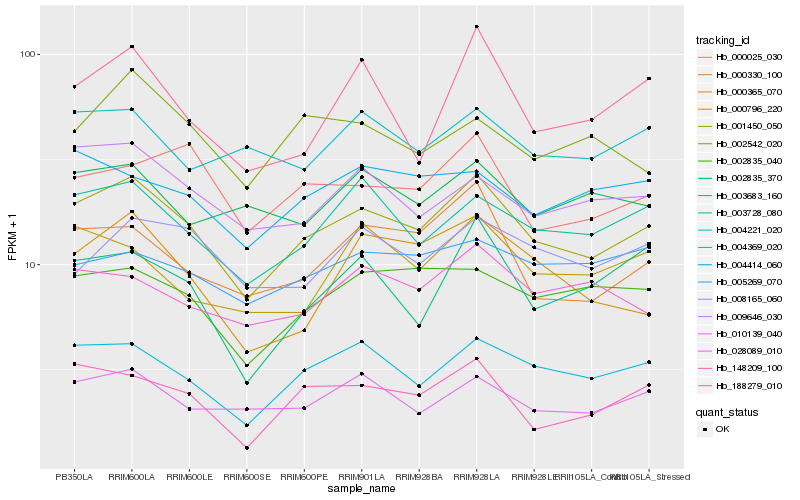
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001450_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004369_020 |
0.0757484988 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 3 |
Hb_003728_080 |
0.0845608286 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like [Jatropha curcas] |
| 4 |
Hb_010139_040 |
0.091782194 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002835_040 |
0.0965865891 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000330_100 |
0.0971156844 |
- |
- |
hypothetical protein VITISV_011650 [Vitis vinifera] |
| 7 |
Hb_148209_100 |
0.0973973553 |
- |
- |
phosphatidylinositol 4-kinase, putative [Ricinus communis] |
| 8 |
Hb_009646_030 |
0.0979846536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000025_030 |
0.103610281 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_028089_010 |
0.1048702497 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |
| 11 |
Hb_008165_060 |
0.1064711992 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 12 |
Hb_004414_060 |
0.1068392769 |
- |
- |
hypothetical protein JCGZ_17225 [Jatropha curcas] |
| 13 |
Hb_005269_070 |
0.1096642019 |
- |
- |
PREDICTED: translation factor GUF1 homolog, organellar chromatophore [Jatropha curcas] |
| 14 |
Hb_004221_020 |
0.109954996 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_188279_010 |
0.1104142466 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC9-like [Tarenaya hassleriana] |
| 16 |
Hb_000796_220 |
0.1111948535 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002835_370 |
0.1114332593 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 18 |
Hb_003683_160 |
0.1123494093 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 19 |
Hb_002542_020 |
0.1137120331 |
- |
- |
PREDICTED: uncharacterized protein LOC105634003 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000365_070 |
0.1138290072 |
- |
- |
PREDICTED: SWR1 complex subunit 2 [Jatropha curcas] |