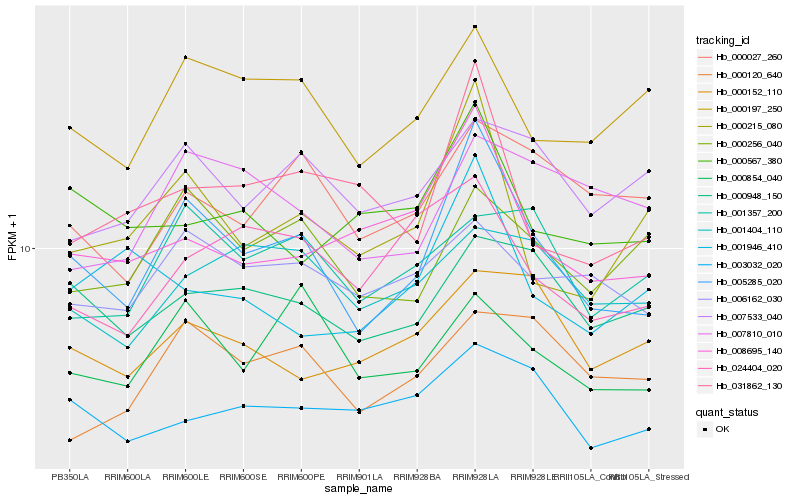
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005285_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000215_080 |
0.1469068524 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008695_140 |
0.1585375341 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 4 |
Hb_031862_130 |
0.1600804039 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000197_250 |
0.1660998399 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 6 |
Hb_000854_040 |
0.169952884 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 7 |
Hb_000948_150 |
0.1756009651 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000256_040 |
0.1757178363 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_006162_030 |
0.1787057289 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X1 [Populus euphratica] |
| 10 |
Hb_024404_020 |
0.1805582826 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 11 |
Hb_003032_020 |
0.1814678884 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55740, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000567_380 |
0.1814824858 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 13 |
Hb_000027_260 |
0.1831169019 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 14 |
Hb_001946_410 |
0.1850061516 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007533_040 |
0.1886689707 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_000152_110 |
0.1918865297 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000120_640 |
0.1922420323 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_007810_010 |
0.1932054785 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001357_200 |
0.1933183383 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_001404_110 |
0.1950305631 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |