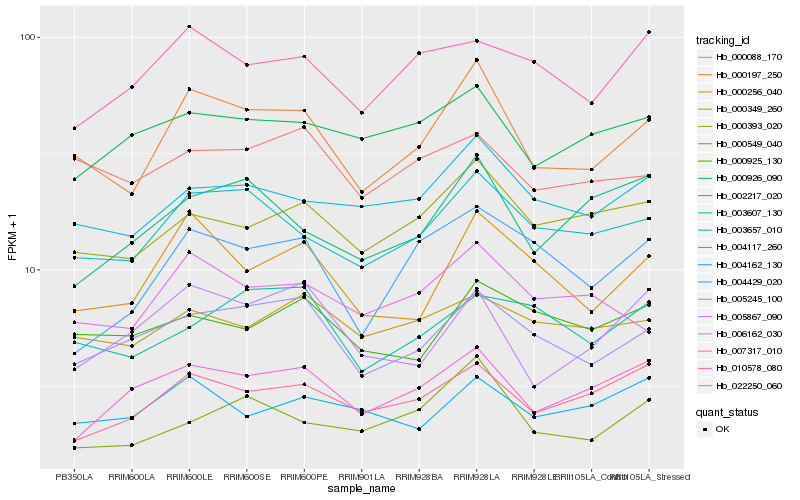
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_250 |
0.0 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 2 |
Hb_003657_010 |
0.0835908575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000256_040 |
0.0933340054 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005245_100 |
0.0958870439 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_004117_260 |
0.0988713455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000393_020 |
0.1025465571 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_006162_030 |
0.1061419919 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X1 [Populus euphratica] |
| 8 |
Hb_022250_060 |
0.1119524932 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000549_040 |
0.1136864101 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007317_010 |
0.1148903492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000088_170 |
0.1152149149 |
- |
- |
PREDICTED: ran-binding protein 10-like [Jatropha curcas] |
| 12 |
Hb_000926_090 |
0.1198382996 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_004429_020 |
0.1198617601 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 14 |
Hb_005867_090 |
0.119897156 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 15 |
Hb_000349_260 |
0.1214567712 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 16 |
Hb_002217_020 |
0.1222345662 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010578_080 |
0.1228797579 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 18 |
Hb_000925_130 |
0.1233741744 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_004162_130 |
0.1241572093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003607_130 |
0.1256132524 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |