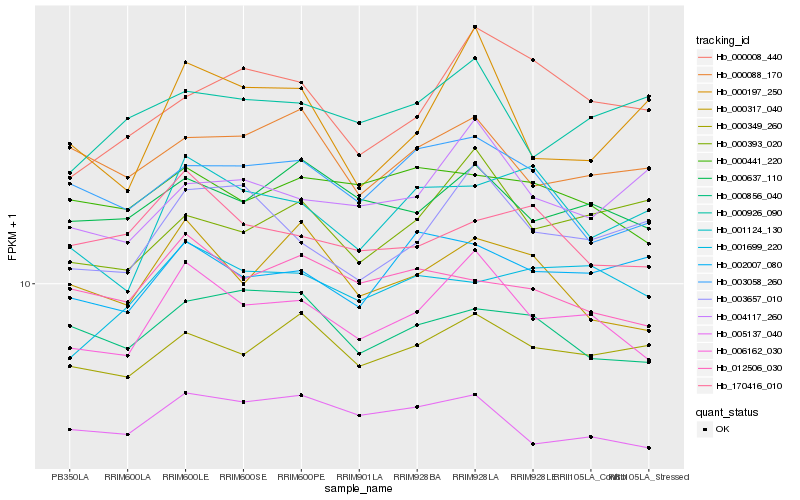
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006162_030 |
0.0 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X1 [Populus euphratica] |
| 2 |
Hb_000637_110 |
0.0860553107 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 3 |
Hb_003657_010 |
0.0864316117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005137_040 |
0.0866828693 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 5 |
Hb_000349_260 |
0.0908913089 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 6 |
Hb_000317_040 |
0.0909440539 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_000393_020 |
0.0991795187 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 8 |
Hb_003058_260 |
0.0999335546 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 9 |
Hb_000441_220 |
0.1005152972 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_012506_030 |
0.1018582916 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 11 |
Hb_000088_170 |
0.1018728596 |
- |
- |
PREDICTED: ran-binding protein 10-like [Jatropha curcas] |
| 12 |
Hb_000926_090 |
0.1022217019 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_004117_260 |
0.1027755067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000856_040 |
0.102949988 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 15 |
Hb_002007_080 |
0.1038928963 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_000008_440 |
0.1040482036 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 17 |
Hb_170416_010 |
0.1051283138 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 18 |
Hb_001699_220 |
0.1057539332 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 19 |
Hb_000197_250 |
0.1061419919 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 20 |
Hb_001124_130 |
0.107233774 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |