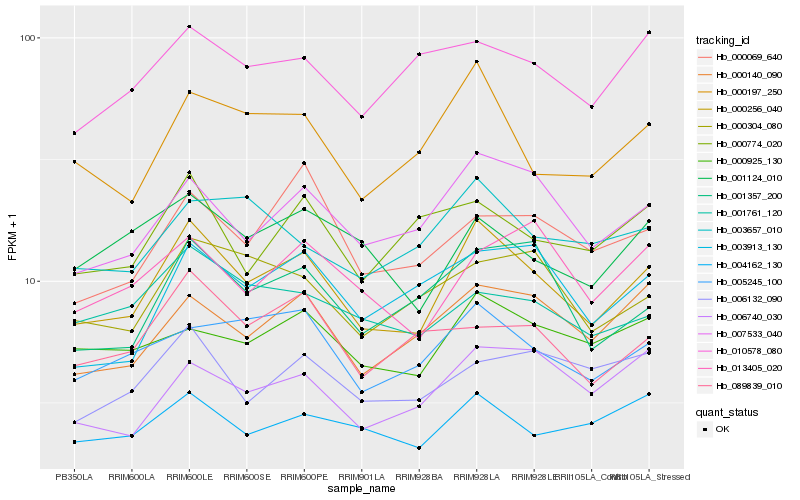
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_040 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_007533_040 |
0.0866934278 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_000197_250 |
0.0933340054 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 4 |
Hb_005245_100 |
0.0993075942 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000140_090 |
0.100769644 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 6 |
Hb_001357_200 |
0.1036891091 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000925_130 |
0.1065448673 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_004162_130 |
0.1078687164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006740_030 |
0.1094994362 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000304_080 |
0.1095162081 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001124_010 |
0.1113335843 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003657_010 |
0.1130272847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003913_130 |
0.1132362506 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 14 |
Hb_000069_640 |
0.1140623108 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 15 |
Hb_001761_120 |
0.1141863611 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 16 |
Hb_000774_020 |
0.114586399 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 17 |
Hb_089839_010 |
0.1148273179 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 18 |
Hb_013405_020 |
0.1153901293 |
- |
- |
- |
| 19 |
Hb_010578_080 |
0.1155722596 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 20 |
Hb_006132_090 |
0.1157564601 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |