| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
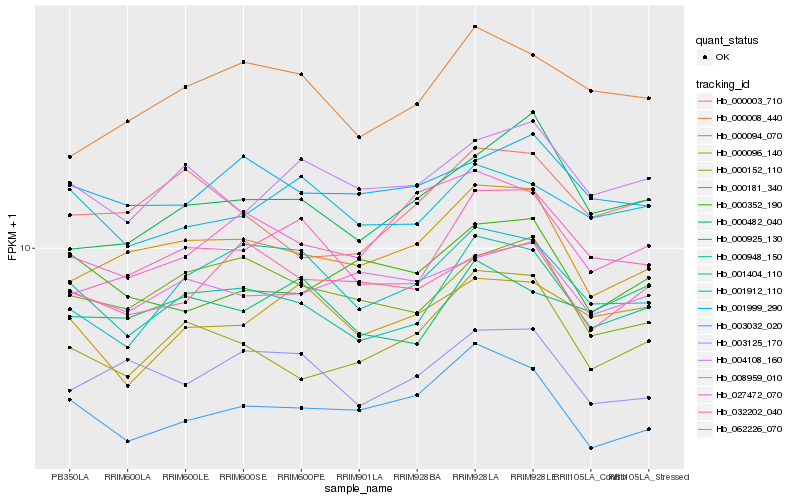
Hb_000948_150 |
0.0 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000152_110 |
0.0910427417 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000181_340 |
0.0924244858 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 4 |
Hb_000094_070 |
0.0948095574 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001912_110 |
0.0975258226 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_004108_160 |
0.100487735 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 7 |
Hb_032202_040 |
0.1007312031 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 8 |
Hb_000003_710 |
0.10149467 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 9 |
Hb_001404_110 |
0.1015958836 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_001999_290 |
0.1039379554 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000008_440 |
0.1107196121 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 12 |
Hb_000925_130 |
0.1109976302 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000352_190 |
0.1120650501 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 14 |
Hb_008959_010 |
0.1121053659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003032_020 |
0.1123328865 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55740, chloroplastic [Jatropha curcas] |
| 16 |
Hb_062226_070 |
0.1126618299 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003125_170 |
0.1136060623 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 18 |
Hb_027472_070 |
0.1147330092 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 19 |
Hb_000096_140 |
0.1149899006 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000482_040 |
0.1156038616 |
- |
- |
Protein YME1, putative [Ricinus communis] |