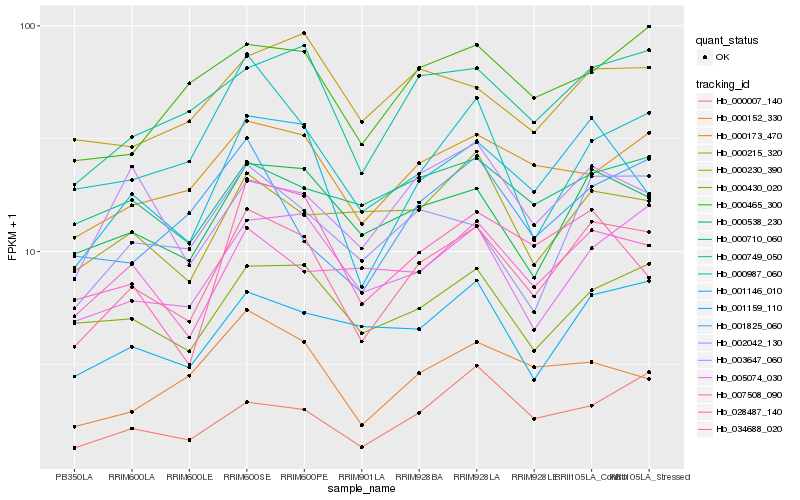
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034688_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 2 |
Hb_001146_010 |
0.0917939312 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 3 |
Hb_000749_050 |
0.0962075918 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 4 |
Hb_000538_230 |
0.1008739595 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005074_030 |
0.1092048406 |
- |
- |
- |
| 6 |
Hb_000430_020 |
0.1092830112 |
- |
- |
- |
| 7 |
Hb_000173_470 |
0.1104425782 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_002042_130 |
0.1123935224 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 9 |
Hb_001159_110 |
0.1156492705 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 10 |
Hb_000465_300 |
0.1201373872 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 11 |
Hb_007508_090 |
0.1240231124 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 12 |
Hb_000710_060 |
0.1279482367 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000007_140 |
0.1280459566 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 14 |
Hb_000215_320 |
0.1304343883 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 15 |
Hb_003647_060 |
0.1312608651 |
- |
- |
PREDICTED: uncharacterized protein LOC105646679 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001825_060 |
0.1313310522 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 17 |
Hb_000152_330 |
0.1315471706 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 18 |
Hb_000230_390 |
0.1326844622 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 19 |
Hb_028487_140 |
0.1327497573 |
- |
- |
PREDICTED: myosin-binding protein 2 [Jatropha curcas] |
| 20 |
Hb_000987_060 |
0.1343975277 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |