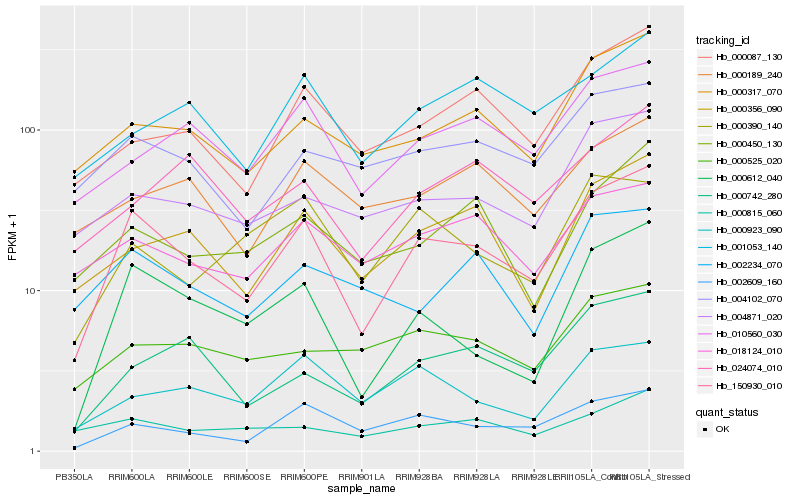
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150930_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000612_040 |
0.1355862502 |
- |
- |
hypothetical protein OsJ_16945 [Oryza sativa Japonica Group] |
| 3 |
Hb_002609_160 |
0.135778178 |
- |
- |
PREDICTED: GDP-mannose transporter GONST2 [Jatropha curcas] |
| 4 |
Hb_000317_070 |
0.1446689456 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 5 |
Hb_000356_090 |
0.1497926258 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 6 |
Hb_010560_030 |
0.1503539436 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 7 |
Hb_000087_130 |
0.1532130477 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 8 |
Hb_004102_070 |
0.1543495111 |
- |
- |
Cytochrome c oxidase subunit 5b-2, mitochondrial -like protein [Gossypium arboreum] |
| 9 |
Hb_004871_020 |
0.1570421518 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 10 |
Hb_000742_280 |
0.1571632492 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_018124_010 |
0.164295794 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001053_140 |
0.1646984395 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000450_130 |
0.1659643356 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 14 |
Hb_000923_090 |
0.1669520984 |
- |
- |
- |
| 15 |
Hb_000390_140 |
0.1695511639 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 16 |
Hb_000525_020 |
0.1699859093 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_002234_070 |
0.170192109 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000189_240 |
0.1703840225 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 19 |
Hb_000815_060 |
0.170864724 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 20 |
Hb_024074_010 |
0.1715111581 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |