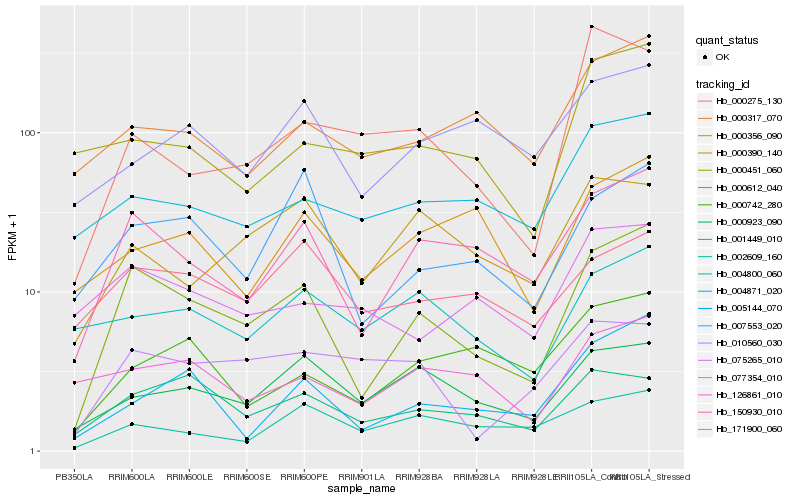
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000612_040 |
0.0 |
- |
- |
hypothetical protein OsJ_16945 [Oryza sativa Japonica Group] |
| 2 |
Hb_150930_010 |
0.1355862502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007553_020 |
0.185747534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005144_070 |
0.1968811667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001449_010 |
0.1999866159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000923_090 |
0.2097348824 |
- |
- |
- |
| 7 |
Hb_004871_020 |
0.2139845546 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 8 |
Hb_000317_070 |
0.214962577 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 9 |
Hb_126861_010 |
0.2171872569 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 10 |
Hb_000390_140 |
0.2182379087 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 11 |
Hb_000742_280 |
0.2194964352 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_171900_060 |
0.2196992868 |
- |
- |
PREDICTED: uncharacterized protein LOC105643368 [Jatropha curcas] |
| 13 |
Hb_077354_010 |
0.2237577263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 14 |
Hb_004800_060 |
0.2251521261 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 15 |
Hb_000356_090 |
0.2252147165 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 16 |
Hb_002609_160 |
0.2257208444 |
- |
- |
PREDICTED: GDP-mannose transporter GONST2 [Jatropha curcas] |
| 17 |
Hb_075265_010 |
0.2272116887 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_000275_130 |
0.2281612281 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |
| 19 |
Hb_000451_060 |
0.2281714696 |
- |
- |
hypothetical protein CICLE_v10010074mg [Citrus clementina] |
| 20 |
Hb_010560_030 |
0.2285569292 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |