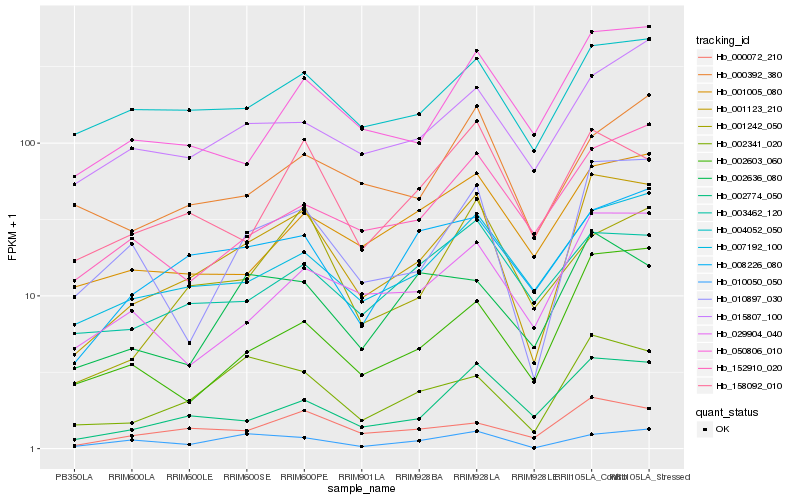
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 2 |
Hb_010897_030 |
0.1353033139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002341_020 |
0.1376539903 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 4 |
Hb_002774_050 |
0.1438593157 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 5 |
Hb_000072_210 |
0.1445596867 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 6 |
Hb_007192_100 |
0.1478893933 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 7 |
Hb_004052_050 |
0.1597285785 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |
| 8 |
Hb_050806_010 |
0.1599759496 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 9 |
Hb_008226_080 |
0.1607836332 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 10 |
Hb_029904_040 |
0.1614225385 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 11 |
Hb_002603_060 |
0.1649205892 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 12 |
Hb_158092_010 |
0.1653301635 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 13 |
Hb_001242_050 |
0.1701592132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001005_080 |
0.1703804601 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_003462_120 |
0.1704651107 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 16 |
Hb_015807_100 |
0.1711309041 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_010050_050 |
0.1718292932 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_002636_080 |
0.1739465655 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 19 |
Hb_152910_020 |
0.1766082857 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 20 |
Hb_000392_380 |
0.1781399931 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |