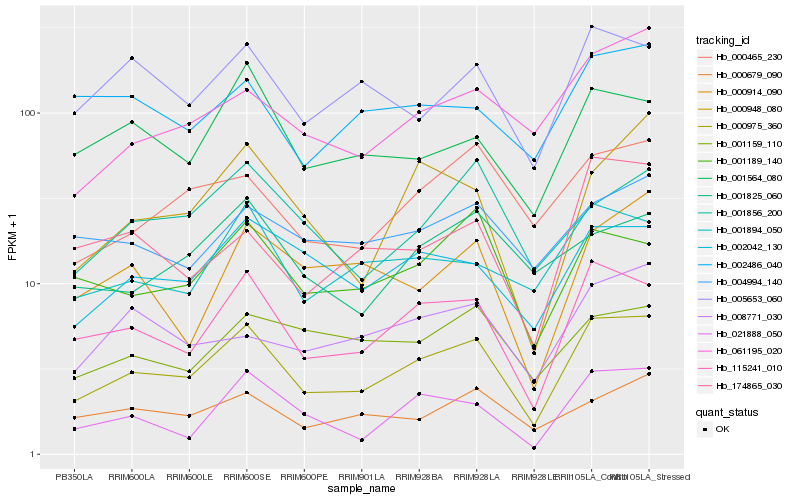
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_360 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_115241_010 |
0.0819542682 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 3 |
Hb_002042_130 |
0.1290852048 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 4 |
Hb_000465_230 |
0.1350578177 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 5 |
Hb_021888_050 |
0.1356371161 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 6 |
Hb_001189_140 |
0.136229805 |
- |
- |
- |
| 7 |
Hb_001894_050 |
0.1363950228 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 8 |
Hb_005653_060 |
0.1374605212 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 9 |
Hb_061195_020 |
0.1396144447 |
- |
- |
PREDICTED: uncharacterized protein LOC101990594 [Microtus ochrogaster] |
| 10 |
Hb_000679_090 |
0.1404569945 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 11 |
Hb_001564_080 |
0.1419950131 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_000948_080 |
0.1436790288 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 13 |
Hb_008771_030 |
0.147851513 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 14 |
Hb_174865_030 |
0.1492284253 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 15 |
Hb_001856_200 |
0.1492976294 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 16 |
Hb_004994_140 |
0.1509539462 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000914_090 |
0.153102391 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 18 |
Hb_001159_110 |
0.1536563263 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 19 |
Hb_001825_060 |
0.1551530761 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 20 |
Hb_002486_040 |
0.1554121248 |
- |
- |
- |