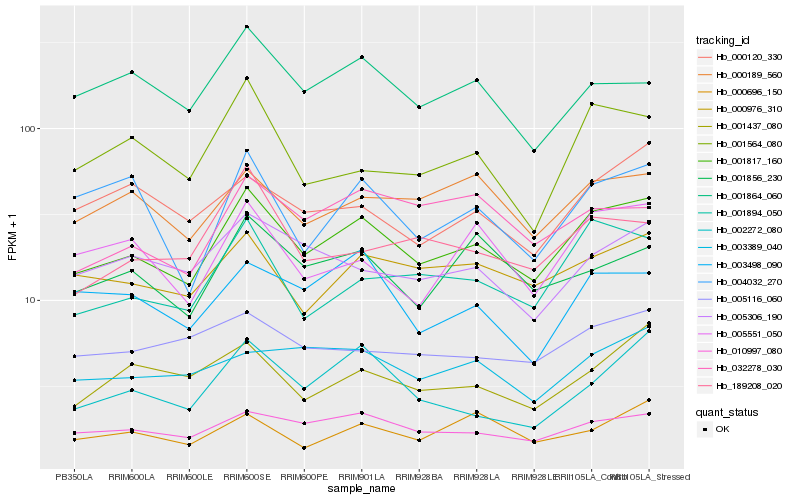
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010997_080 |
0.0970083758 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000976_310 |
0.1010755761 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001894_050 |
0.1035731952 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 5 |
Hb_000189_560 |
0.1124017082 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 6 |
Hb_005306_190 |
0.1160709474 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 7 |
Hb_001856_230 |
0.1172582199 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 8 |
Hb_005551_050 |
0.1175017896 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001437_080 |
0.1193877028 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 10 |
Hb_005116_060 |
0.1198133254 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003498_090 |
0.1199459579 |
- |
- |
hypothetical protein RCOM_0867850 [Ricinus communis] |
| 12 |
Hb_001864_060 |
0.1217538345 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 13 |
Hb_032278_030 |
0.1221676926 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_003389_040 |
0.1223591798 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 15 |
Hb_001564_080 |
0.1227503298 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_189208_020 |
0.1228490031 |
- |
- |
LRX2, putative [Ricinus communis] |
| 17 |
Hb_002272_080 |
0.1239024282 |
- |
- |
- |
| 18 |
Hb_000696_150 |
0.1240302729 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g51320 [Jatropha curcas] |
| 19 |
Hb_000120_330 |
0.1245637534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004032_270 |
0.124581427 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |