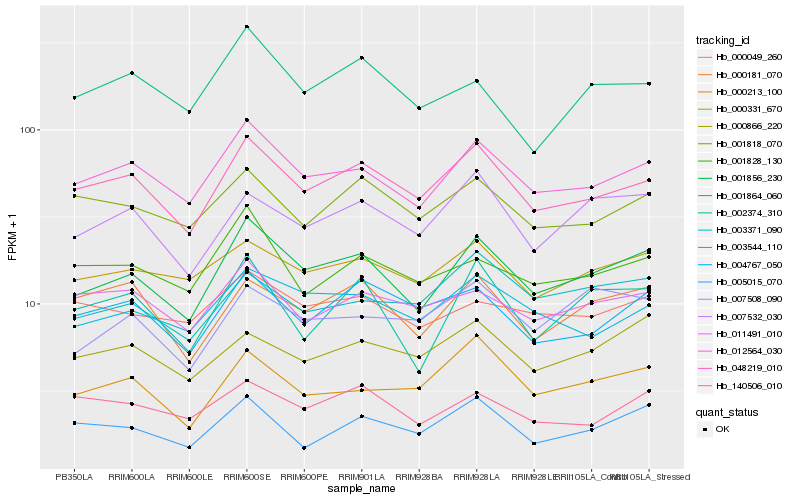
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_230 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 2 |
Hb_012564_030 |
0.0600200008 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 3 |
Hb_048219_010 |
0.0720646093 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 4 |
Hb_003544_110 |
0.0930681591 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007532_030 |
0.0946984603 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 6 |
Hb_000331_670 |
0.0955293066 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 7 |
Hb_002374_310 |
0.097694109 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 8 |
Hb_000181_070 |
0.1013376289 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_004767_050 |
0.1026450118 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 10 |
Hb_001828_130 |
0.1037958764 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 11 |
Hb_000213_100 |
0.1038353483 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 12 |
Hb_001818_070 |
0.1066610891 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 13 |
Hb_001864_060 |
0.1070378685 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 14 |
Hb_000866_220 |
0.1072879763 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 15 |
Hb_000049_260 |
0.1074936388 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_007508_090 |
0.1077215624 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_003371_090 |
0.1079465032 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 18 |
Hb_005015_070 |
0.1081059688 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 19 |
Hb_140506_010 |
0.109059914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170-like [Jatropha curcas] |
| 20 |
Hb_011491_010 |
0.1093702356 |
- |
- |
Protein YME1, putative [Ricinus communis] |