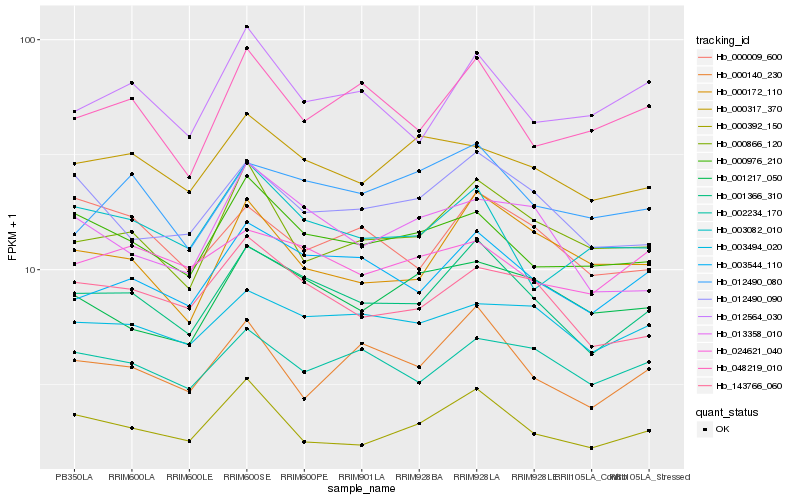
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_310 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 2 |
Hb_003544_110 |
0.0692413196 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_143766_060 |
0.0803601401 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 4 |
Hb_012490_090 |
0.0883856767 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 5 |
Hb_000976_210 |
0.0887541981 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003494_020 |
0.0910346433 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 7 |
Hb_000392_150 |
0.0942216261 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g74580 [Jatropha curcas] |
| 8 |
Hb_012490_080 |
0.0950824319 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 9 |
Hb_000866_120 |
0.095737694 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 10 |
Hb_013358_010 |
0.0960565218 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 11 |
Hb_002234_170 |
0.0964198147 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000172_110 |
0.0966863594 |
- |
- |
- |
| 13 |
Hb_000317_370 |
0.0968125759 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 14 |
Hb_001217_050 |
0.0979285313 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 15 |
Hb_000140_230 |
0.0994069578 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_048219_010 |
0.0994372151 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 17 |
Hb_003082_010 |
0.1011029866 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 18 |
Hb_000009_600 |
0.1012893095 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 19 |
Hb_012564_030 |
0.1020234324 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 20 |
Hb_024621_040 |
0.1025517881 |
transcription factor |
TF Family: ERF |
hypothetical protein L484_014666 [Morus notabilis] |