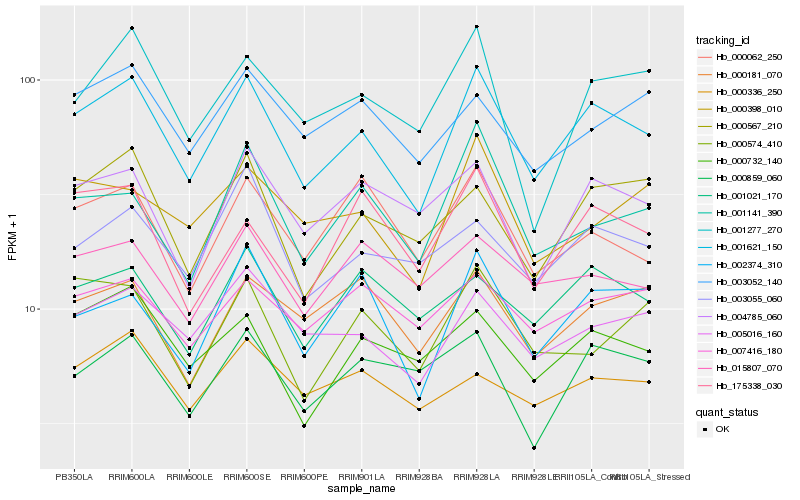
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_150 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 2 |
Hb_005016_160 |
0.0937776901 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 3 |
Hb_002374_310 |
0.0981498231 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 4 |
Hb_000732_140 |
0.1009559965 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC [Jatropha curcas] |
| 5 |
Hb_000567_210 |
0.1016793945 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2-like [Jatropha curcas] |
| 6 |
Hb_001021_170 |
0.1039269326 |
- |
- |
PREDICTED: uncharacterized protein LOC105642474 [Jatropha curcas] |
| 7 |
Hb_004785_060 |
0.1059640893 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 8 |
Hb_001141_390 |
0.1063535871 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 9 |
Hb_001277_270 |
0.1066042534 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_175338_030 |
0.1077919781 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 11 |
Hb_003055_060 |
0.1081966822 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000062_250 |
0.1082063512 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 13 |
Hb_000398_010 |
0.1095218162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003052_140 |
0.1096994398 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 15 |
Hb_007416_180 |
0.1114561798 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 16 |
Hb_000859_060 |
0.1116626409 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 17 |
Hb_000181_070 |
0.1117038597 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000336_250 |
0.1119725996 |
transcription factor |
TF Family: mTERF |
hypothetical protein B456_002G027700 [Gossypium raimondii] |
| 19 |
Hb_015807_070 |
0.1127253337 |
- |
- |
PREDICTED: autophagy-related protein 18f [Jatropha curcas] |
| 20 |
Hb_000574_410 |
0.1132729898 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600 [Jatropha curcas] |