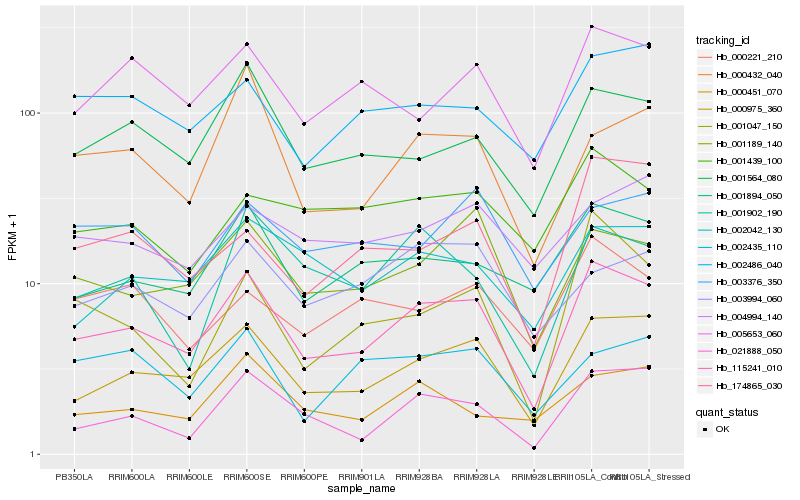
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115241_010 |
0.0 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 2 |
Hb_000975_360 |
0.0819542682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001564_080 |
0.1259003072 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_001894_050 |
0.1274114946 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 5 |
Hb_001189_140 |
0.130868857 |
- |
- |
- |
| 6 |
Hb_021888_050 |
0.1311841943 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 7 |
Hb_005653_060 |
0.1382565753 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 8 |
Hb_002042_130 |
0.143093085 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 9 |
Hb_000432_040 |
0.1468893024 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 10 |
Hb_001902_190 |
0.1485864583 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 11 |
Hb_002486_040 |
0.1490697263 |
- |
- |
- |
| 12 |
Hb_000451_070 |
0.152410814 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_174865_030 |
0.1538210034 |
- |
- |
PREDICTED: hemiasterlin resistant protein 1 [Eucalyptus grandis] |
| 14 |
Hb_001047_150 |
0.1541489436 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 15 |
Hb_000221_210 |
0.1551939826 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_001439_100 |
0.1573517783 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 17 |
Hb_003994_060 |
0.1588647085 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 18 |
Hb_004994_140 |
0.1618167686 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003376_350 |
0.1626619822 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002435_110 |
0.162685891 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |